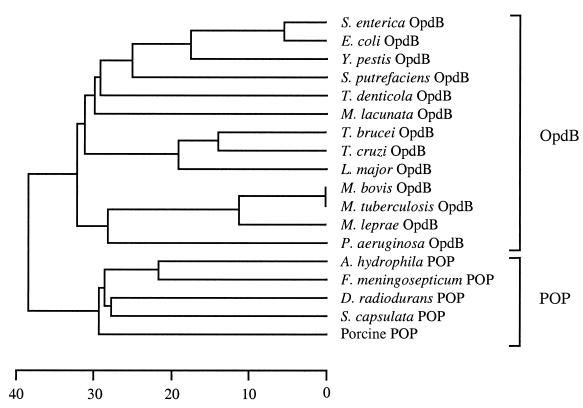
FIG. 1.
Phylogenetic relationship of members of the POP family of serine peptidases. An unrooted dendrogram was prepared by comparing the full-length amino acid sequences of 18 members of the POP family using the CLUSTAL W alignment software of the MEGALIGN program (DNASTAR) with a PAM250 weight table set with the following parameters: ktuple = 1, gap penalty = 3, gap window = 5. The scale at the bottom measures the distance between sequences. The units indicate the number of substitution events (as a percentage). Sequences were obtained from the GenBank/EBI database under the following accession numbers: D10976 (E. coli OpdB), AF078916 (T. brucei OpdB), U69897 (T. cruzi OpdB), AF109875 (L. major OpdB), D38405 (M. lacunata OpdB), Z80226 (Mycobacterium tuberculosis OpdB), Z95151 (Mycobacterium leprae OpdB), AAK39550 (T. denticola OpdB), D14005 (Aeromonas hydrophila POP), D10980 (Flavobacterium meningosepticum POP), U08343 (Deinococcus radiodurans POP), AB010298 (Sphingomonas capsulata POP) and M64227 (porcine brain POP). The OpdB sequences for Y. pestis and Mycobacterium bovis were extracted from their partially sequenced genomes at the Sequencing Group at the Sanger Centre (available at www.sanger.ac.uk). The OpdB sequences from S. putrefaciens and Vibrio cholerae were extracted from their partially sequenced genomes at The Institute for Genome Research (available at www.tigr.org). The Pseudomonas OpdB sequence was extracted from the partially sequenced Pseudomonas genome at the Pseudomonas Genome Project (available at www.pseudomonas.com).