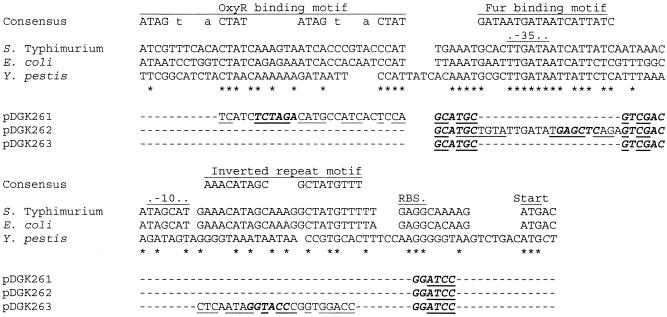
FIG. 1.
Wild-type and mutant candidate cis-acting motifs in the mntH promoter. Proximal regions of the mntH promoters from S. enterica serovar Typhimurium (24), E. coli (24, 27), and Y. pestis (5) were aligned and compared to consensus motifs for OxyR (40) and Fur (2, 9). The S. enterica serovar Typhimurium and E. coli mntH promoters both have a candidate OxyR-binding site upstream of the predicted −35 region and a candidate Fur-binding site overlapping and extending downstream of the −35 region. Also highlighted is an inverted-repeat motif between the predicted −10 region and the ribosome binding site (RBS) of the mntH coding region. Of these three motifs, only a putative Fur-binding site is readily discernible in the Y. pestis mntH promoter. Below these motifs are lines indicating the substitutions made in the three cis-acting S. enterica serovar Typhimurium promoter mutants used in this study, pDGK261, pDGK262, and pDGK263. Identical nucleotides in the three promoter regions (asterisks), novel restriction sites introduced between motifs or within block substitutions (described in Materials and Methods) (bases in bold italic type), and isopositional sequences from the bacteriophage P22 Pant promoter (bases in lightface roman type) are indicated. As some substituted bases fortuitously matched those found in the wild-type S. enterica serovar Typhimurium mntH promoter, the bases actually changed in pDGK261, pDGK262, and pDGK263 are underlined. In the consensus OxyR-binding motif the bases shown in lowercase type are common but not completely conserved bases at these sites (38).