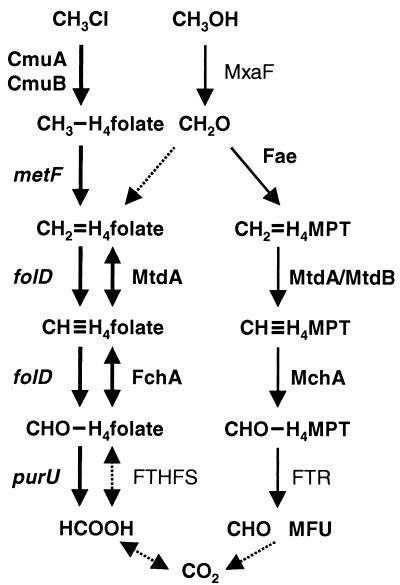
FIG. 1.
Proposed model of pterin-dependent C1 metabolism in M. chloromethanicum CM4. Transformations shown by light arrows indicate reactions involved in growth of M. extorquens AM1 with methanol (6). Transformations shown by heavy arrows are proposed to be specific to the chloromethane-degrading strain M. chloromethanicum CM4. CmuA, chloromethane-corrinoid methyltransferase (34); CmuB, methylcobalamin-H4folate methyltransferase (35); metF, methylene-H4folate reductase; folD, bifunctional methylene-H4folate dehydrogenase-methenyl-H4folate cyclohydrolase; purU, 10-formyl-H4folate hydrolase; MtdA, NADP-dependent methylene-H4folate-H4MPT dehydrogenase (39); FchA, methenyl-H4folate cyclohydrolase; (30); FTHFS, formyl-H4folate synthetase; Fae, formaldehyde-activating enzyme (41); MtdB, NAD(P)-dependent methylene-H4MPT dehydrogenase (10); Mch, methenyl-H4MPT cyclohydrolase; (30) FTR, formylmethanofuran-H4MPT formyltransferase (29). Enzymes indicated in boldface were detected in strain CM4 at the DNA level (reference 13 and PCR data not shown).