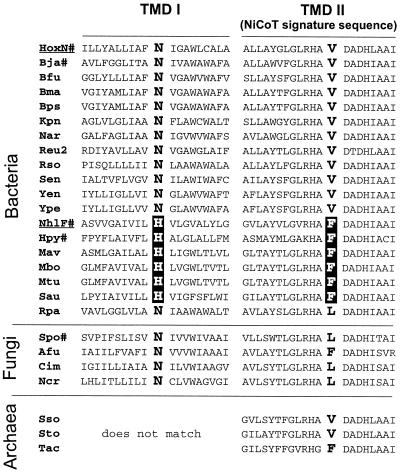
FIG. 4.
Sequence alignment of putative TMDs I and II of NiCoTs from bacteria, archaea, and fungi. The list contains experimentally investigated permeases (#), sequences from completed genome projects, and preliminary genomic and expressed sequence tag sequences. HoxN from R. eutropha and NhlF from R. rhodochrous are underlined. The correlation between an Asn or His residue in TMD I and a Val, Phe, or Leu residue within the NiCoT signature sequence in TMD II is highlighted. The similarity between TMD I of the archaeal transporters and their bacterial and fungal counterparts is not obvious. The program CLUSTALW (http://clustalw.genome.ad.jp) was used for multiple alignments. Afu, A. fumigatus; Bfu, Burkholderia fungorum; Bja, Bradyrhizobium japonicum (11); Bma, Burkholderia mallei; Bps, Burkholderia pseudomallei; Cim, C. immitis; Hpy, NixA of H. pylori (12, 13); Kpn, Klebsiella pneumoniae; Mav, Mycobacterium avium; Mbo, Mycobacterium bovis; Mtu, Mycobacterium tuberculosis; Ncr, N. crassa; Nar, Novosphingobium aromaticivorans; Reu2, R. eutropha; Rso, Ralstonia solanacearum; Rpa, R. palustris; Sen, Salmonella enterica; Spo, S. pombe (6); Sso, S. solfataricus; Sto, S. tokadaii; Tac, T. acidophilum; Yen, Yersinia enterocolitica; Ype, Yersinia pestis.