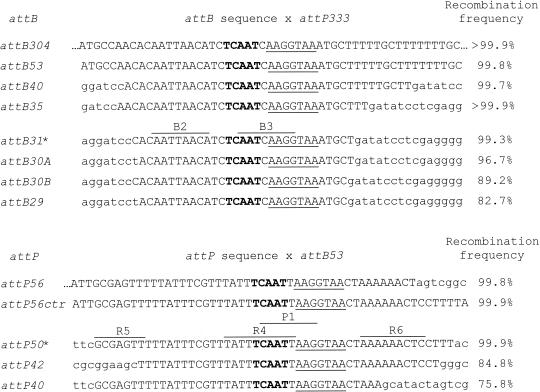
FIG. 2.
Efficiency of integrative recombination mediated by TP901-1 integrase between att sites of various sizes in E. coli. For each sequence shown, the uppercase letters represent att DNA sequences, while the lowercase letters represent flanking vector sequences. Boldface type indicates the 5-bp common core, and the underlined sequence is the 7-bp identical region shared by attB and attP. Previously identified repeats are indicated by lines above the attB31 and attP50 sequences, as discussed in the text. The asterisks next to these two att sites indicate that they are the smallest sites that were still fully active in this assay. The recombination frequency is the intramolecular integration frequency, calculated by determining the ratio of white colonies to total colonies and multiplying by 100. Each att site is followed by a number indicating the length of the att DNA sequence. In attB30A and attB30B there were deletions from the left and right sides of the sequence, respectively. attP56ctr is an attP sequence in which the att base pairs are centered precisely around the 5-bp core. Each attB was tested by integrative recombination against a 333-bp attP, while each attP was tested against a 53-bp attB. Frequency calculations were made by using bacterial strain DH-TPInt and are based on total numbers of bacterial colonies ranging from 500 to 8,800.