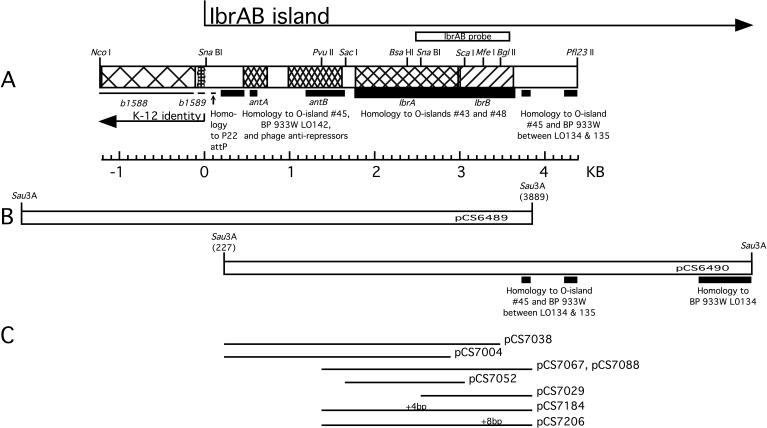
FIG. 1.
Map of the IbrAB island. (A) Gene names and the junction of the IbrAB island with the E. coli K-12 chromosome are indicated below the map; ORFs ibrA and ibrB are translated from left to right. Coordinate 0 marks the end of the K-12 identity. Regions of E. coli O157:H7 homology to O islands no. 43 and 48 (strain EDL933) and SpLE1 (Sakai strain) are indicated by thick lines; homology to bacteriophage 933W and O island no. 45 of E. coli O157:H7 EDL933 is indicated by lines of intermediate thickness, and regions identical to K-12 are indicated by thin lines below the map; the position of the IbrAB probe is shown above. BP, bacteriophage. (B) Primary plasmid clones. Nucleotide coordinates within the IbrAB island of the sequenced end of each primary clone are shown in parentheses. (C) Plasmids containing deletions and insertions affecting ibrA or ibrB. The positions of frameshifting insertions are indicated as +4 bp (at BsaHI) and +8 bp (at MfeI), respectively, above plasmids pCS7184 and pCS7207.