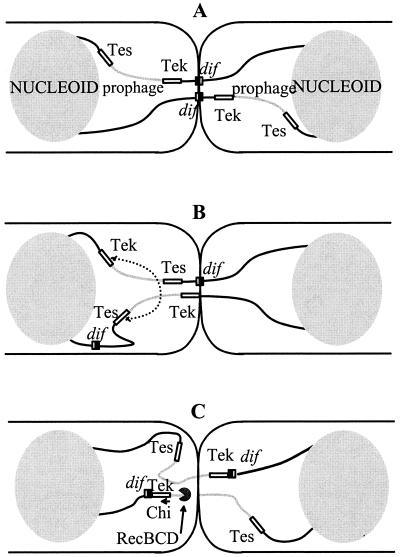
FIG. 3.
Model for prophage excision and recovery of circular chromosome monomers from a dimer in a xerC mutant. (A) The strain harbors an iso-oriented prophage in the terminus region, with no associated perturbation of the positioning of dif sites at division. Trapping by the septum immobilizes the dif regions and leads to generation of double-strand ends. The absence of overlap leaves no possibility for recombination to yield a circular monomer in either of the daughter cells. Both cells are doomed. (B) A pseudo-DAZ generated at the junction of an antioriented prophage and the chromosome can result in septal trapping of DNA distant from dif, whereas the positioning of the second thread is correct. The resulting overlap of terminus sequences can be used to generate a prophage-cured circular monomer by homologous recombination between the indicated Tes and Tek elements in the cell to the left, while the cell to the right is doomed. (C) When the prophage maps at or near dif, positioning operates to place prophage DNA under the septum. In the example depicted (chosen for simplicity), breakage in the trapped zone results in simultaneous prophage excision and chromosome repair, provided the RecBCD enzyme, responsible for the repair, encounters the stimulating Chi site present in the tet sequence. Both cells can recover a viable monomer chromosome.