Abstract
The origin of replication (oriC) region in some clinical strains of Mycobacterium tuberculosis is a hot spot for IS6110 elements. To understand how clinical strains with insertions in oriC can replicate their DNA, we characterized the oriC regions of some clinical strains. Using a plasmid-based oriC-dependent replication assay, we showed that IS6110 insertions that disrupted the DnaA box sequence CCGTTCACA abolished oriC activity in M. tuberculosis. Furthermore, by using a surface plasmon resonance technique we showed that purified M. tuberculosis DnaA protein binds native but not mutant DnaA box sequence, suggesting that stable interactions of the DnaA protein with the CCGTTCACA DnaA box are crucial for replication of oriC plasmids in vivo. Replacement by homologous recombination of the CCGTTCACA DnaA box sequence of the laboratory strain M. tuberculosis H37Ra with a mutant sequence did not result in nonviability. Together, these results suggest that M. tuberculosis strains have evolved mechanisms to tolerate mutations in the oriC region and that functional requirements for M. tuberculosis oriC replication are different for chromosomes and plasmids.
Mycobacterium tuberculosis, the causative agent of tuberculosis, has infected more than one-third of the world's population and accounts for 3,000,000 deaths each year. M. tuberculosis grows slowly, with a doubling time of about 24 h. The genetic and biochemical factors that are responsible for the slow growth rate of M. tuberculosis are unknown. Earlier studies designed to understand the replication initiation process in M. tuberculosis revealed that the dnaA-dnaN intergenic region functions as oriC (17). The corresponding region in other species of mycobacteria has also been shown to function as oriC in native hosts (10, 17, 19, 20). The oriC region is A-T rich and contains several putative DnaA boxes, defined as sequences with one to three mismatches to the consensus sequence TT (G/C) TCC ACA (17). Deletions in the oriC region abolish oriC activity (17, 18), and point mutations in the DnaA box sequences of Mycobacterium smegmatis oriC severely decreased oriC activity (18), indicating the importance of both the integrity of oriC and the sequence of the DnaA boxes in mycobacterial replication initiation.
The M. tuberculosis H37Rv genome contains 16 copies of IS6110 and several other insertion sequences (5). Recent IS6110 sequence mapping data for clinical strains of M. tuberculosis revealed that the dnaA-dnaN intergenic region is a hot spot for IS6110 insertion (7). While many of these insertions were located outside the DnaA boxes, some disrupted one putative DnaA box with the sequence CCGTTCACA. Interestingly, no other DnaA boxes in the oriC of M. tuberculosis were found to be disrupted by IS6110 sequences. The IS6110 insertion in the DnaA box was designated as A-4 (8). The deletion and point mutation data of the mycobacterial oriC (17, 18) raise questions as to whether the CCGTTCACA DnaA box is essential for oriC replication and, if so, how clinical strains of M. tuberculosis with IS6110 insertions in oriC initiate chromosomal DNA replication. To get insights into the replication initiation process in M. tuberculosis, we have characterized the oriC regions from selected clinical strains and asked two specific questions: (i) do mutations (insertions) in the oriC region of clinical strains interfere with the autonomous replication activity of respective oriC plasmids? (ii) Are mutations in the DnaA box of the oriC region tolerated on the chromosome of a laboratory strain of M. tuberculosis? Our results indicate that mutations in the A-4 DnaA box which abolish replication of plasmids are tolerated on the chromosome, suggesting that functional requirements for M. tuberculosis oriC are different for chromosomes and plasmids.
MATERIALS AND METHODS
Strains.
Escherichia coli Top10 (Invitrogen) and a laboratory strain of M. tuberculosis H37Ra were used in the present study. Middlebrook 7H9 medium supplemented with oleic acid-dextrose-sodium chloride-catalase was used to culture M. tuberculosis strains. When required, transformants were selected on agar plates supplemented with kanamycin (20 μg/ml), sucrose (0.2%), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 50 μg/ml). Genomic DNA prepared from the following clinical strains of M. tuberculosis was used to amplify the oriC region: TN670 and TN6333 (A-1 insertion), TN5110 (A-4 insertion), and TN3990 (A-10 insertion). TN670 and TN6333 are two different but related M. tuberculosis strains (8). The IS6110 mapping analyses revealed that the two M. tuberculosis strains could have diverged recently following the unique insertion of IS6110 in oriC (8).
Transformation experiments. (i) oriC plasmids containing IS6110 insertions.
OriC activity was defined as the ability of a DNA fragment from M. tuberculosis to render autonomous replication to E. coli plasmids in an M. tuberculosis host (10, 17, 18). DNA fragments containing the dnaA-dnaN intergenic region from clinical strains were amplified by PCR, cloned into pZErO 2.1 vector (Invitrogen), and used in transformation experiments. These DNA fragments were 814 bp in length and contained IS6110 sequences either outside the DnaA boxes at two different locations designated as A-1 (pMQ387 and pMQ388) and A-10 (pMQ371) or inside the CCGTTCACA DnaA box (pMQ373). As a control, a pMQ219 plasmid construct carrying a corresponding 814-bp DNA fragment from M. tuberculosis H37Rv was used (17).
(ii) oriC containing a BglII mutation in the DnaA box (pJor2).
A BglII sequence in the CCGTTCACA DnaA box at the A-4 site was created by using a PCR mutagenesis protocol (25) with the pMQ219 plasmid as a template. The oligonucleotide primers A (AGATCTAACGGAACCGCCGGGACTG), which binds to the DnaA box and its upstream sequence at the A-4 site, and B (AGATCTACAACCCACGCCTCATCC), which binds to the oriC sequence downstream of the A-4 insertion site, were used. The BglII site is underlined, and nucleotide sequence complementary to the oriC sequence is shown in boldface type. Restriction digestion of the amplified product with BglII followed by self-ligation resulted in the production of a mutant pMQ219 plasmid containing BglII sequence at the A-4 insertion site.
(iii) oriC containing mutations in the IS6110 sequence.
Two plasmids designated pQ373D and pQM373D and containing mutations in different regions of the IS6110 sequence were constructed. Digestion of pMQ373 with SmaI and ScaI enzymes followed by self-ligation produced pQ373D, which lacked an approximately 900-bp internal DNA fragment of the IS6110 sequence. A derivative of this plasmid lacking the left inverted repeat (IR) was constructed by PCR using primer A (see above) and C (AGATCTCAGTTCTTGGAAAGGATGGG), which binds to the IS6110 sequence downstream of the left IR. The BglII site is underlined and the nucleotide sequence complementary to the IS6110 sequence is shown in boldface. PCR amplification and restriction digestion of the amplified product with BglII followed by self-ligation resulted in the production of a recombinant plasmid, pQM373D, lacking the left IR. The nucleotide sequence of the oriC region was verified in both constructs to ensure that no mutations were created during the plasmid construction.
Recombination experiments.
The chromosomal oriC sequence was replaced with a mutant oriC sequence by homologous recombination, following a two-step strategy (16). First, suicidal recombination delivery vectors carrying mutant oriC sequences were transformed into M. tuberculosis to produce a single-crossover (SCO) strain. Next, double crossovers (DCOs) were selected to yield mutant strains. Two suicidal recombination delivery vectors containing mutant oriC sequences, designated pJOR4 and pJOR14 and each containing different lengths of homologous sequences flanking the BglII site, were constructed. For the construction of pJOR4, an 814-bp oriC DNA fragment containing the BglII mutation in the A-4 DnaA box was cloned into p2NIL, a nonreplicating mycobacterial vector (16). In the next step, a 6.1-kb PacI marker gene cassette carrying the genes lacZ (for blue color), aph (responsible for resistance to kanamycin), and sacB (responsible for sensitivity to sucrose) was released from a pGOAL17 vector (16) and cloned into the above vector. This construct contained approximately 300 and 500 bp of homologous DNA flanking the 5′ and 3′ ends of the BglII site, respectively. The pJOR14 vector was constructed by PCR using a 5-kb M. tuberculosis dnaA region (Fig. 1) as a template. First, a 2,060-bp 5′ DNA fragment upstream of the A-4 site was amplified using the primers 5′-AACTGCAGCCCGGCAACCGCTTCAGGGC-3′ and 5′-GAAGATCTGAACGGAACCGCCG-GGACTG-3′. Similarly, the 1,360-bp 3′ DNA fragment downstream of the A-4 site was amplified with primers 5′-GCGGATCCGTCCAACATCATCGGCACCCG-3′ and 5′-GAAGATCTACAACCCACGCCTCATCC-3′. The BamHI, PstI, and BglII restriction endonuclease sites used to facilitate cloning are underlined. In the next step, a 1.3-kb DNA fragment containing the 3′ flanking sequence was released by BglII-NdeI digestion and fused with the 2-kb 5′ flanking region at the BglII site. The 3.4-kb DNA fragment containing the BglII mutation in the A-4 site was cloned as a BamHI-PstI fragment into p2NIL, and the selection cassette from pGOAL17 was inserted as described above (see Fig. 4). The SCOs were selected on agar plates containing kanamycin and X-Gal and their sensitivity to sucrose was confirmed. To promote selection of DCO events, SCOs were streaked on antibiotic-free plates, resuspended in broth, and spread on agar plates containing sucrose and X-Gal. Potential DCOs were confirmed as white colonies that were sucrose resistant and kanamycin sensitive. The presence of the BglII mutation was verified by restriction digestion of genomic DNA followed by Southern hybridization.
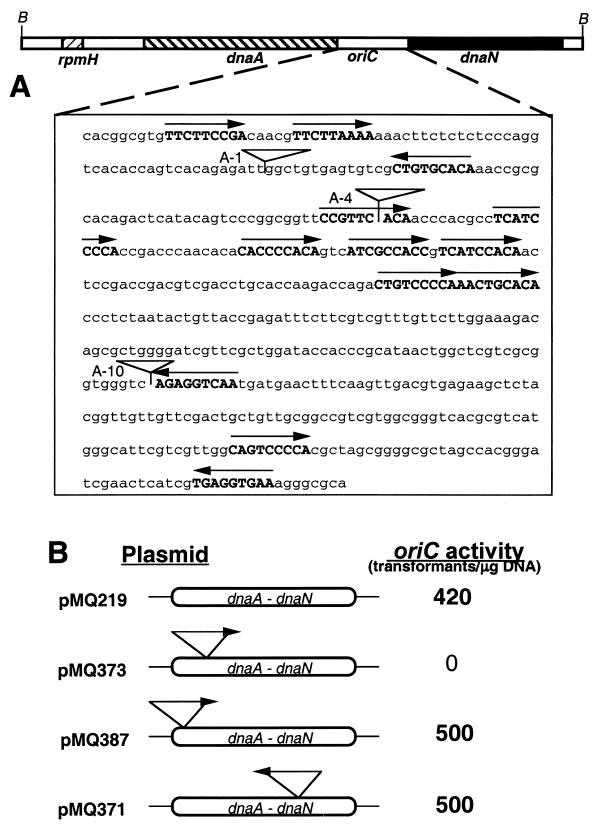
FIG. 1.
oriC activity of the dnaA-dnaN intergenic region containing IS6110. (A) Organization of the dnaA-dnaN intergenic region. A 5-kb BamHI DNA fragment carrying the M. tuberculosis dnaA region (not drawn to scale) and the sequence of the dnaA-dnaN intergenic region are shown. B, BamHI site. DnaA boxes are shown in bold capital letters, and the arrows show the orientation of the DnaA boxes. Inverted triangles indicate the location of the IS6110 insertion sequences at the A-1, A-4, and A-10 sites. (B) oriC activity of plasmids containing IS6110 sequences. Plasmids containing oriC sequences derived from the clinical strains were transformed into the laboratory strain M. tuberculosis H37Ra, and the ability of the plasmids to transform and replicate as stable autonomous sequences was determined. The oriC activity is shown as the total number of transformants obtained per microgram of input DNA. pMQ219 is the control oriC plasmid derived from M. tuberculosis H37Rv (17). It includes an 814-bp DNA fragment containing the dnaA-dnaN intergenic region and its flanking regions. Although not shown, the oriC activity of pMQ388 is similar to that of pMQ387.
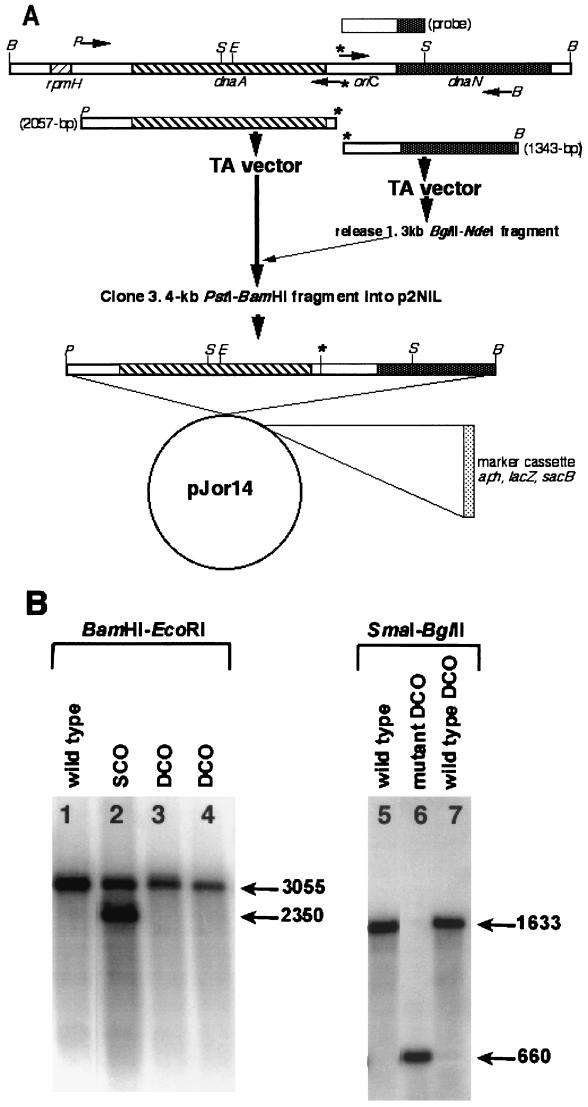
FIG. 4.
Replacement of the wild-type oriC sequence with mutant sequence. (A) Construction of pJOR14 recombination delivery vector. A 5-kb M. tuberculosis dnaA region, containing the rpmH, dnaA, and dnaN genes, and oriC are shown. The locations of genes are shown as different hatched boxes. The sites of restriction endonuclease enzymes are denoted by single letter codes as follows: B, BamHI; S, SmaI; P, PstI; E, EcoRI. The locations of primers used to amplify 2- and 1.3-kb DNA fragments are shown as arrowheads. The star above the arrowhead indicates the BglII site incorporated to create a mutation in the A-4 DnaA box. The amplification products along with their sizes are shown below the dnaA region. The DNA probe used in Southern hybridization experiments is shown above the dnaA region. The 3.4-kb DNA BamHI-PstI fragment was cloned into p2NIL, and a 6.1-kb PacI marker cassette was inserted to create pJOR14 vector. This vector was used to generate SCOs and, subsequently, DCOs. (B) Southern analyses of the DCOs. Genomic DNA of mycobacterial strains (mutant and wild-type DCOs) was digested with the different enzymes shown, transferred to nitrocellulose membranes, and hybridized with a 600-bp SmaI-BglII fragment. Note that the sizes of the SCO products with BamHI-EcoRI enzymes are different. Both wild-type and mutant DCOs were distinguished by BglII-SmaI digestion.
Surface plasmon resonance (SPR) experiments and DnaA protein.
Binding of M. tuberculosis DnaA protein to oligonucleotides containing DnaA boxes was examined using a BIAcore X instrument. The recombinant M. tuberculosis DnaA protein was purified as a His-DnaA fusion protein on nickel affinity columns as described previously (27). A biotinylated oligonucleotide with a single DnaA box flanked on either side by its native oriC sequence (5′-CGGCGGTT-CCGTTCACAACCCACGC-3′) was synthesized, annealed to complementary oligonucleotide, and used to immobilize the streptavidin-coated sensor chip. The DnaA box sequence is shown in bold letters. The CCGTTCACA DnaA box disrupted with the BglII sequence at the same location as the A-4 insertion (5′-CGGCGGTTCCGTTCAGATCTACAAC-3′), was also synthesized, annealed to complementary strand, and used as a control. The BglII site is underlined. In addition, oligonucleotide with a scrambled sequence (5′-AAGTAAGTATATAGTTTAAGTAAGT-3′) was synthesized and used as a second control. Biotinylated oligonucleotides were captured on streptavidin-coated SA sensor chips, different concentrations of DnaA protein were injected over the sensor chip surface, and both association and dissociation were recorded.
RESULTS AND DISCUSSION
IS6110 insertions located outside the DnaA boxes do not affect oriC activity.
We showed earlier that an 814-bp M. tuberculosis oriC DNA fragment containing the 527-bp dnaA-dnaN intergenic region, 177 bp of the 3′ end of the dnaA gene, and 110 bp of the 5′ end of the dnaN gene functioned as oriC in M. tuberculosis hosts. Furthermore, we showed that plasmids containing less than 814 bp of oriC DNA were unstable, whereas those containing more than 814 bp of oriC DNA, i.e., the dnaA-dnaN intergenic region and different lengths of the upstream dnaA gene including the rpmH-dnaA intergenic region, did not increase the transformation efficiency of the oriC plasmids (17). We also found that plasmids containing the 5-kb M. tuberculosis dnaA region, i.e., the rpmH, dnaA, and dnaN genes and their intergenic regions, exhibited oriC activity (17). However, the majority of the recovered plasmids contained deletions in the rpmH-dnaA intergenic region and the dnaA-coding region. Presumably, the rpmH-dnaA intergenic region may interfere with oriC activity in plasmid context. To test whether DNA fragments derived from clinical strains containing IS6110 insertions in the dnaA-dnaN intergenic region support oriC activity, appropriate plasmid constructs were transformed into the laboratory strain of M. tuberculosis. Plasmid constructs bearing IS6110 insertions outside the DnaA box sequences designated A-1 (pMQ387 and pMQ388) and A-10 (pMQ371) supported oriC activity like that of the control plasmids without insertions (pMQ219; Fig. 1), but the construct designated A-4 (pMQ373) did not (Fig. 1). The transformation efficiency of these positive plasmids was comparable to that of the control (≈0.4 × 104 transformants/μg of input plasmid DNA).
To investigate if the observed oriC activities of the pMQ371, pMQ387, and pMQ388 plasmids are due to accumulation of compensatory mutations in the oriC region or due to changes in the IS6110 site of insertion following transformation and growth, plasmids from the stable transformants were isolated and DNA was analyzed using several restriction enzymes. In addition, the nucleotide sequences of the entire 814-bp oriC DNA fragment (minus the IS6110 sequence) of the input as well as the recovered plasmids were determined. Both the restriction digestion patterns and nucleotide sequence of the oriC region, including the sites of insertion of IS6110 of the input as well as the recovered plasmids, were found to be identical (data not shown). Together, these results suggest that the presence of IS6110 insertions outside the DnaA box, i.e., A-1 and A-10, do not affect oriC activity, whereas those in the DnaA box do. Presumably, the integrity of the DnaA box is important for replication of oriC plasmids.
The integrity of the CCGTTCACA DnaA box is important for oriC activity.
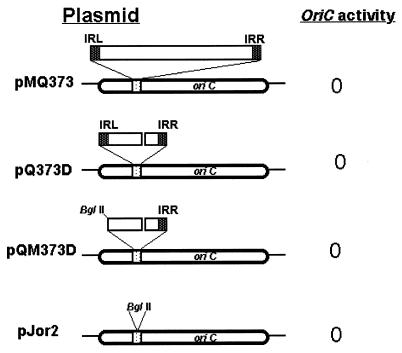
The IS6110 sequence belongs to the IS3 family of insertion sequences and contains 28 bp of IR flanking the open reading frame for transposase, which recognizes and processes the IR for mobility and transposition (11). Transposition events involving IS6110 and other repetitive elements have been suggested to furnish the pathogen with an important mechanism for altering gene expression (24). We considered a possibility that the lack of oriC activity of pMQ373 plasmids is not due to the disruption of the CCGTTCACA sequence but due to some deleterious activity originating from the IS6110 sequences. To address this question, plasmid constructs containing mutations in the IS6110 sequences (pQ373D and pQM373D; Fig. 2) were transformed into M. tuberculosis strain H37Ra. It is expected that mutations that remove potential promoter and IS6110 internal sequence would minimize the IS6110 activity (11). Transformation of the M. tuberculosis strain with the above recombinant plasmids failed to produce any viable transformants, suggesting that the lack of plasmid replication is not due to insertion sequence activity affecting the biology of the DnaA box. Next, to investigate whether sequences other than the IS6110 in the CCGTTCACA DnaA box at the A-4 site are tolerated, we examined the oriC activity of the pJor2 plasmid containing the BglII restriction enzyme sequence at the A-4 site. Consistent with disruption of the DnaA box by IS6110, the BglII insertion containing the oriC plasmid, designated pJor2, did not exhibit oriC activity (Fig. 2). Together, these results suggest that integrity of the CCGTTCACA DnaA box is important for oriC replication, and mutations or insertions that affect the integrity of the CCGTTCACA DnaA box abolish oriC activity.
FIG. 2.
oriC activity of pMQ373 derivatives. oriC plasmids with different mutations are shown to the left and the oriC activity is shown to the right. Only the CCGTTCACA (DnaA box) sequence is boxed for clarity. The IS6110 sequence is shown as an inverted triangle above the oriC sequence. The gap in IS6110 represents the internal deletion. The left IR (IRL) and right IR (IRR) are marked as black squares flanking IS6110.
The DnaA protein is not proficient in interacting with the mutant DnaA box.
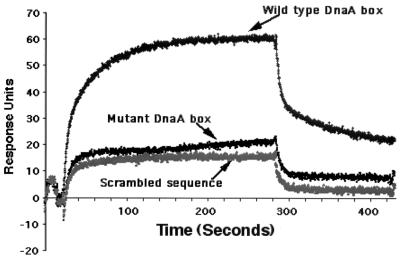
Replication is believed to be initiated when the DnaA protein recognizes and binds to the DnaA boxes located in the oriC region. The interaction of DnaA with the DnaA boxes is believed to be the first step in triggering the initiation process (7, 15). The lack of replication of pMQ373, pQM373D, pQ373D, and pJor2 plasmids in M. tuberculosis hosts could be due to the lack of binding of the DnaA protein to the mutant DnaA boxes. It should be noted that although sequence comparisons identified approximately 13 DnaA box-like sequences in the oriC region of M. tuberculosis, actual binding of M. tuberculosis DnaA protein to any of these boxes has not been confirmed (17). To test whether CCGTTCACA is a DnaA box and to test the above prediction, the dnaA gene from M. tuberculosis H37Rv was cloned and expressed in E. coli from the bacteriophage T7 promoter, and the recombinant protein was purified on nickel affinity columns as a His-DnaA fusion protein (27). Preliminary results indicated that the recombinant protein hydrolyzed ATP, and it bound and reduced the mobility of the oriC DNA in agarose gels (27). The ability of the recombinant protein to interact with the CCGTTCACA DnaA box was investigated by SPR using a BIAcore X instrument. Change in mass at the surface of a sensor chip is measured by SPR (4, 13). The major advantage of this technique is that protein-DNA interactions are monitored in real time. Injection of the DnaA protein (125 nM) to the wild-type DnaA box sensor chip surface resulted in a rapid binding within 25 s which then slowed and reached equilibrium by 100 s (Fig. 3). This was followed by a slow dissociation. Further washing with the buffer resulted in only a slow decay of the signal. From these results we infer that CCGTTCACA is an authentic DnaA box. At a similar protein concentration (125 nM; Fig. 3) and even at twofold-higher concentrations (data not shown), the DnaA protein did not stably associate with the mutant DnaA box. The binding profiles of the DnaA to the mutant DnaA box were comparable to that of the control scrambled sequence.
FIG. 3.
DnaA protein binding to the DnaA box. Biotinylated oligonucleotides containing either the wild-type DnaA box sequence CCGTTCACA or the mutant sequence CCGTTCagatctACA or a scrambled sequence (see text) were coupled to the streptavidin sensor chip surface. DnaA protein at different concentrations was incubated for 4 min at room temperature in 50 mM Tris acetate buffer (pH 8.0), 0.5 mM magnesium acetate, 0.3 mM EDTA, 10 mM ammonium chloride, with 0.005% Tween 20, 1 mM ATP, and 30 ng of poly(dA-dT) competitor DNA/ml and then injected over the sensor chip surface at a flow rate of 5 μl/min. Total injection time was approximately 5 min. The sensor chip was washed with the above buffer 3 min after the injection to record dissociation. Only results obtained with 125 nM DnaA protein are shown. For the sake of clarity, spikes in the beginning and the end of the injection were removed.
The lack of binding of DnaA protein to the scrambled and mutant DnaA box sequences in vitro and the lack of replication of plasmids containing the mutant DnaA box sequences in vivo suggest that CCGTTCACA is an important DnaA box and that stable interactions of the DnaA protein with this DnaA box are crucial for replication of these plasmids in vivo. The oriC sequence (minus the IS6110 sequence) of the clinical strains is essentially identical to that of the corresponding sequence of the laboratory strain of M. tuberculosis, suggesting that the replication initiation mechanisms are similar in these strains. Since mutations (insertions) in one DnaA box abolished replication of the plasmids in M. tuberculosis hosts, similar mutations in the oriC region of the clinical strains would be expected to be lethal. In contrast, the clinical strains tolerate the IS6110 sequences in the oriC region. These results are different from those observed with E. coli oriC plasmids (6, 9). E. coli oriC has five DnaA boxes that are distributed throughout oriC. E. coli DnaA protein does not interact with any of the mutant DnaA boxes in vitro, although point mutations in DnaA boxes either individually or in combination are tolerated in vivo (1, 6, 9). These results demonstrate that binding of E. coli DnaA protein to mutant DnaA boxes is possible in vivo (9). Presumably, such binding involves cooperativity. Recently, Speck et al. (23) showed that the E. coli DnaA protein bound cooperatively to mutant DnaA boxes following its binding to adjacent wild-type DnaA boxes. Their results suggested that cooperative DnaA protein interactions are important for oriC activity (14, 23).
M. tuberculosis oriC, unlike its E. coli counterpart, contains 13 DnaA box-like sequences which are distributed in the entire oriC (17). Furthermore, the sequences of the designated DnaA boxes of M. tuberculosis oriC are clearly different from those of the E. coli boxes (17). We showed that the M. tuberculosis DnaA protein bound to one DnaA box (Fig. 3). It is unknown whether M. tuberculosis DnaA binds to all designated DnaA boxes and whether cooperativity is important for M. tuberculosis oriC function. Since cooperativity appears to be a common feature of all the DnaA proteins that have been investigated (12, 14, 21-23), we think that the M. tuberculosis DnaA-oriC interactions would also involve cooperativity, and such cooperative interactions involving all DnaA boxes are critical for the formation of an effective oriC initiation complex. Presumably, nonoptimal binding of M. tuberculosis DnaA protein to the mutant DnaA box in vivo could result in a weak cooperativity. This in turn could lead to the formation of a defective initiation complex, thereby affecting the replication of oriC plasmids in vivo. Further experiments are required to characterize cooperativity.
oriC plasmid constructs containing the rpmH-dnaA intergenic region do not support oriC activity.
The replication of DNA containing IS6110 insertions in the oriC region of the clinical strains suggests that the clinical strains have evolved mechanisms to tolerate IS6110 insertions in the DnaA boxes located on the chromosome. One possibility is that the clinical strains with IS6110 insertions in their oriC use alternate replication origins for initiation of chromosomal DNA replication. The rpmH-dnaA intergenic region of either M. tuberculosis or Mycobacterium avium does not function as oriC in the respective native hosts (10, 17). Both regions are approximately 65% similar. The rpmH-dnaA intergenic region of M. avium, however, functions as oriC in M. tuberculosis hosts. These results suggested that the rpmH-dnaA intergenic region is a silent replication origin and its activity could be unmasked in the absence of the functional primary oriC, i.e., the dnaA-dnaN intergenic region (10). Assuming that the M. tuberculosis dnaA-dnaN intergenic region containing the IS6110 insertions in oriC is defective for replication initiation, then one possibility is that the rpmH-dnaA intergenic region or possibly other sequences on the chromosome promote replication of DNA.
To test whether the rpmH-dnaA intergenic region functions as oriC, two series of experiments were carried out. First, oriC activities of plasmid constructs containing a 5-kb rpmH-dnaA-dnaN DNA fragment with a BglII mutation in the CCGTTCACA DnaA box at the A-4 locus in M. tuberculosis hosts were examined. Transformation of M. tuberculosis strain H37Ra with mutant oriC plasmids did not result in any viable transformants, whereas plasmids containing the entire dnaA region did (∼0.4 × 104 transformants/μg of input plasmid DNA), as previously reported (17). These results are not surprising, because the intact CCGTTCACA DnaA box sequence is essential for replication of oriC plasmids (Fig. 1 and 2), and the M. tuberculosis rpmH-dnaA intergenic region in a wild-type background does not support oriC activity (10, 17). Second, to overcome incompatibility problems, if any, that are associated with oriC, the ability of the M. tuberculosis rpmH-dnaA intergenic region to function as oriC in an M. avium host was examined. These experiments did not result in any viable transformants, presumably because of the poor transformation efficiencies of M. avium strains (∼1 to 10 transformants per μg of input DNA), which are approximately 2 to 3 orders of magnitude less than that of M. tuberculosis strains (10).
Mutations in the A-4 DnaA box on the chromosome are tolerated in the laboratory strain of M. tuberculosis.
In the case of E. coli it has been shown that the DnaA box R4, which is essential for replication of oriC plasmids, is dispensable for replication at the chromosomal oriC (1, 2). Deletion of the R4 DnaA box, however, led to asynchronous initiation of chromosomal replication. More recently, Weigel et al. (26) characterized the chromosomal oriC by replacing it with different mutated oriC sequences. Their results showed that origins with mutations in the R1 box are nonfunctional whereas those in R2, R3, and R4 are, suggesting that the functionalities of these mutated oriC are greater on the chromosome than on a minichromosome (26). Presumably, this may be the case with the oriC of the clinical strains of M. tuberculosis. To test whether this feature is unique to clinical strains or if other strains of M. tuberculosis can tolerate mutations in the CCGTTCACA DnaA box of oriC, we attempted to replace the chromosomal oriC of M. tuberculosis H37Ra with a mutant oriC containing the BglII mutation in the CCGTTCACA box (pJOR4), using homologous recombination following the two-step gene replacement protocol (16). If an M. tuberculosis strain with a mutated oriC survives, this would indicate that mutations in the CCGTTCACA DnaA box of oriC on the chromosome are tolerated in the laboratory strain as well. Electroporation of M. tuberculosis H37Ra with the pJOR4 construct did not result in any viable SCO recombinants. Presumably, the length of homology used, i.e., 300 and 500 bp of homologous DNA to the 5′ and 3′ ends of the BglII mutation, respectively, in the pJOR4 construct, was not sufficient to promote recombination events.
To promote selection of SCO recombinants, we constructed a pJOR14 recombination delivery vector containing 2 kb upstream and 1.4 kb downstream of homologous sequences flanking the BglII mutation (Fig. 4A ). Electroporation of this vector into M. tuberculosis produced two SCOs that were blue, resistant to kanamycin, and sensitive to sucrose. One SCO, designated RGM43, was further processed to select DCO strains that were white, sensitive to kanamycin, and resistant to sucrose. PCR amplification followed by restriction digestion with the BglII enzyme of the oriC region of 10 potential DCOs revealed wild-type and mutant DCO patterns in a 3:7 ratio (data not shown; see below). To further confirm these results, genomic DNA of a potential mutant (RGM46) and wild-type (RGM47) DCO was digested with different enzymes and analyzed by Southern hybridization (Fig. 4B). Digestion with BamHI-EcoRI enzymes identified DNA bands corresponding to either a 3-kb fragment for wild type (Fig. 4B, lane 1), both 2.3- and 3-kb fragments for SCO (Fig. 4B, lane 2), and a 3-kb fragment for both wild-type (Fig. 4B, lane 3) and mutant (Fig. 4B, lane 4) DCOs. Since a 6-bp BglII insertion sequence is not expected to change the size of the DNA band in agarose gels, the fragment sizes obtained from either wild-type or mutant DCOs are of the same size. Digestion of the genomic DNA of the RGM46 and RGM47 strains with SmaI-BglII enzymes followed by Southern hybridization confirmed the presence of a BglII mutation in the A-4 site of oriC in the RGM46 DCO (Fig. 4B, compare lane 6 with lanes 7 and 5). In broth, the RGM46 mutant DCO grew like that of RGM47 and the parent strain, with little or no difference in the doubling time (data not shown).
Together, these results indicate that mutations in the A-4 DnaA box of oriC, which abolish replication of plasmids, are tolerated on the chromosome. Presumably, the functional requirements for replication of oriC plasmids and chromosomes are also different in M. tuberculosis, much like the results reported with E. coli oriC systems (1-3, 26). Thus, survival of the clinical strains with IS6110 insertions in oriC, i.e., the dnaA-dnaN intergenic region, is due to their ability to tolerate these mutations. The IS6110 insertions in the oriC region of M. tuberculosis strains are naturally occurring mutations (8). In this regard, it is pertinent that some clinical strains of M. tuberculosis contain two IS6110 insertions with up to 300-bp deletions in the oriC region (e.g., M. tuberculosis TN6278 [B. N. Kreiswirth and N. Kurepina, unpublished data]). The deleted region spans the DNA beginning at the A-1 site to the end of A-10 and includes a total of nine presumptive DnaA boxes (see Fig. 1A for locations of DnaA boxes). Further characterization of the oriC regions in the clinical strains will enable us to understand how these large deletions in oriC are also tolerated in M. tuberculosis.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health to M.M. (AI41406) and M.R. (AI48417). J.D. was supported by a Fulbright fellowship during the study period.
We thank Bonnie Plikaytis, Centers for Disease Control and Prevention, Atlanta, Ga., for preliminary oriC sequence information on M. tuberculosis strain TN6278, Bhavna Gordhan for details on gene replacement protocols, and Mark Atkinson for his interest and support of the work.
REFERENCES
- 1.Asai, T., D. B. Bates, E. Boye, and T. Kogoma. 1998. Are minichromosomes valid model systems for DNA replication control? Lessons learned from Escherichia coli. Mol. Microbiol. 29:671-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, D. B., T. Asai, Y. Cao, M. W. Chambers, G. W. Cadwell, E. Boye, and T. Kogoma. 1995. The DnaA box R4 in the minimal oriC is dispensable for initiation of Escherichia coli chromosome replication. Nucleic Acids Res. 23:3119-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, D. B., E. Boye, T. Asai, and T. Kogoma. 1997. The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:12497-12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bondeson, K., A. Frostell-Karlsson, L. Fagerstam, and G. Magnusson. 1993. Lactose repressor-operator DNA interactions: kinetic analyses by a surface plasmon resonance biosensor Anal. Biochem. 214:245-251. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Holz, A., C. Schaefer, H. Gille, W. R. Jueterbock, and W. Messer. 1992. Mutations in the DnaA binding sites of the replication origin of Escherichia coli. Mol. Gen. Genet. 233:81-88. [DOI] [PubMed] [Google Scholar]
- 7.Kornberg, A., and T. Baker. 1991. DNA replication. W.H. Freeman and Company, New York, N.Y.
- 8.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Sooligen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 9.Langer, U., S. Richter, A. Roth, C. Weigel, and W. Messer. 1996. A comprehensive set of DnaA-box mutations in the replication origin, oriC, of Escherichia coli. Mol. Microbiol. 21:301-311. [DOI] [PubMed] [Google Scholar]
- 10.Madiraju, M. V. V. S., M. H. Qin, K. Yamamoto, M. A. Atkinson, and M. Rajagopalan. 1999. The dnaA gene region of Mycobacterium avium and the autonomous replication activities of its 5′ and 3′ flanking regions. Microbiology 145:2913-2921. [DOI] [PubMed] [Google Scholar]
- 11.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majka, J., J. Zakrzewska-Czerwinska, and W. Messer. 2001. Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces. J. Biol. Chem. 276:6243-6252. [DOI] [PubMed] [Google Scholar]
- 13.Malmqvist, M. 1993. Biospecific interaction analysis using biosensor technology. Nature 361:186-187. [DOI] [PubMed] [Google Scholar]
- 14.Messer, W., F. Blaesing, D. Jakimowicz, M. Krause, J. Majka, J. Nardmann, S. Schaper, H. Seitz, C. Speck, C. Weigel, G. Wegrzyn, M. Welzeck, and J. Zakrzewska-Czerwinska. 2001. Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading. Biochimie 83:5-12. [DOI] [PubMed] [Google Scholar]
- 15.Messer, W., H. Hartmann-Kuhlein, U. Langer, E. Mahlow, A. Roth, S. Schaper, B. Urmoneit, and B. Woelker. 1992. The complex for replication initiation of Escherichia coli. Chromosoma 102:1-6. [DOI] [PubMed] [Google Scholar]
- 16.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 17.Qin, M. H., M. V. V. S. Madiraju, and M. Rajagopalan. 1999. Characterization of the functional replication origin of Mycobacterium tuberculosis. Gene 233:121-130. [DOI] [PubMed] [Google Scholar]
- 18.Qin, M. H., M. V. V. S. Madiraju, S. Zachariah, and M. Rajagopalan. 1997. Characterization of the oriC region of Mycobacterium smegmatis. J. Bacteriol. 179:6311-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajagopalan, M., M. H. Qin, D. R. Nash, and M. V. V. S. Madiraju. 1995. Mycobacterium smegmatis DnaA region and autonomous replication activity. J. Bacteriol. 177:6527-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar, L., H. Fsihi, E. de Rossi, G. Riccardi, C. Rios, S. T. Cole, and H. E. Takiff. 1996. Organization of the origins of replication of the chromosomes of Mycobacterium smegmatis, Mycobacterium leprae and Mycobacterium tuberculosis and isolation of a functional origin from M. smegmatis. Mol. Microbiol. 20:283-293. [DOI] [PubMed] [Google Scholar]
- 21.Schaper, S., and W. Messer. 1995. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270:17622-17626. [DOI] [PubMed] [Google Scholar]
- 22.Speck, C., and W. Messer. 2001. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speck, C., C. Weigel, and W. Messer. 1999. ATP- and ADP-DnaA protein, a molecular switch in gene regulation. EMBO J. 18:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stemmer, C., and S. K. Morris. 1992. Enzymatic inverse PCR: a restriction site independent, single-fragment method for high efficiency, site directed mutagenesis. BioTechniques 13:214-220. [PubMed] [Google Scholar]
- 26.Weigel, C., W. Messer, S. Preiss, M. Welzeck, and E. Boye. 2001. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40:498-507. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto, K., S. Muniruzzaman, M. Rajagopalan, and M. V. V. S. Madiraju. 2002. Modulation of the Mycobacterium tuberculosis DnaA protein interactions with adenine nucleotides by acidic phospholipids. Biochem. J. 363:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]