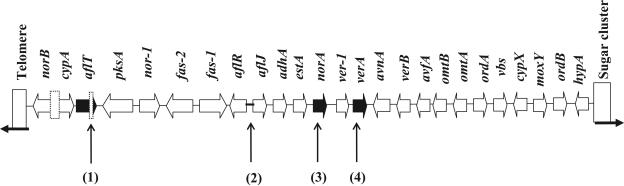
FIG. 1.
Structure of the aflatoxin biosynthesis gene homolog cluster in Aspergillus oryzae RIB 40. Boldface black arrows indicate genes encoding deduced polypeptides with less than 93% amino acid similarity to those of A. flavus (arrow length is not proportional to gene size). Dotted boxes indicate deletions. The boldface line indicates mutations in a recognized consensus sequence within a promoter region. The numbered vertical arrows indicate specific mutations: 1, a 257-bp deletion resulting in loss of 1 of 14 putative transmembrane regions; 2, base substitutions in consensus sequences for putative AreA and FacB binding sites; 3, a frameshift mutation resulting in a truncation; 4, amino acid substitutions.