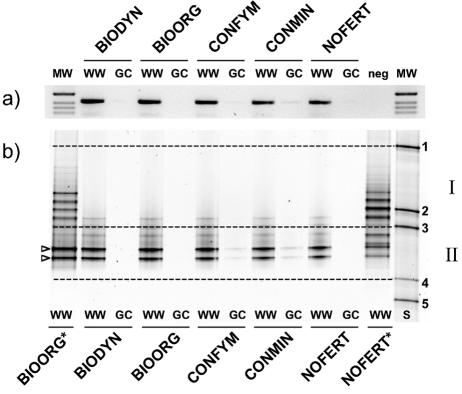
FIG. 4.
PCR (a) and DGGE (b) analyses of CRP SSU rRNA gene fragments amplified from bulk soil DNA extracts. Analyzed samples included five different soils from the DOK field experiment (lanes BIODYN, BIOORG, CONFYM, CONMIN, and NOFERT) planted with winter wheat (lanes WW) or grass-clover (lanes GC). For PCR analysis, a 1-kb marker (lane MW) was used as a DNA size standard, and a negative control (neg) was included. The size of the CRP PCR products was 417 bp. The migration standard (S) and migration regions (I and II) for DGGE analysis are described in the legend to Fig. 2. Asterisks designate community DGGE patterns from soils and were included for direct comparison to the data shown in Fig. 2. Open arrowheads, dominant bands detected in all winter wheat soils.