Abstract
Horizontal gene transfer (HGT) is generally considered a possible mechanism by which bacteria acquire new genetic properties. Especially when pathogenicity genes are involved, HGT might have important consequences for humans. In this report we describe a case study of HGT in which a transcriptional activator, ComK of Bacillus subtilis, was introduced into a heterologous host species, Lactococcus lactis. ComK is the central regulator of competence development, activating transcription by binding to a ComK-binding site, a so-called K-box. Interestingly, L. lactis does not contain a comK gene, but it does contain almost 400 putatively functional K-boxes, as well as homologues of a number of competence genes. In this study, the effect of HGT of B. subtilis comK into L. lactis was investigated by determining the effects on the transcription profile using DNA microarray analyses. Production of wild-type ComK was shown to stimulate the transcription of 89 genes and decrease the expression of 114 genes. Notably, potential direct effects (i.e., genes preceded by a K-box) were found mainly among repressed genes, suggesting that ComK functions as a repressor in L. lactis. This is a remarkable difference between L. lactis and B. subtilis, in which ComK almost exclusively activates transcription. Additional DNA microarray analyses with a transcription activation-deficient but DNA-binding ComK variant, ComKΔC25, demonstrated that there were similar effects on gene regulation with this variant and with wild-type ComK, confirming that the direct effects of ComK result from interference with normal transcription through binding to available K-boxes. This study demonstrates that horizontal gene transfer can have dramatic effects that are very different than those that are expected on basis of the original functionality of a gene.
Bacteria are subjected to rapid evolution by continuous changes in their genome sequences, resulting in new genotypes. Natural selection causes survival of the fittest and elimination of really disadvantageous genotypes. Occasionally, new genotypes are so advantageous that they become dominant and spread over the population. The most common causes of genomic changes involve only the cells' own genome, brought about by alterations at the nucleotide level (e.g., introduction of mutations in the DNA due to damage provoked by UV light, chemical agents, DNA polymerase mistakes, etc.) or alterations of larger parts of the genome (e.g., duplications, repetitions, or strand inversions). In addition, new genetic abilities can be acquired by horizontal gene transfer (HGT), for which three main mechanisms have been reported: (i) transformation of naturally competent bacteria (e.g., Bacillus subtilis) by the uptake of environmental DNA, (ii) transduction of DNA by bacteriophage infection, and (iii) conjugation (transfer of DNA between adjacent bacteria by pilin structures) (5). Several studies have suggested that specific genes have been acquired by HGT, including the ermG gene found in Bacillus sphaericus and seven intestinal Bacteroides species (36), the cap operon, which is thought to have been acquired by Staphylococcus epidermidis by plasmid transfer from Bacillus anthracis (11), and the ycdB gene, which has recently been exchanged between lactococci and enterobacteria (4; for a review see reference 31). Often, the presumptions are based on differences in the G+C contents of the genes and the rest of the genome, suggesting that there were different origins (9). Especially when pathogenic bacteria or acquired antibiotic resistance is involved, HGT challenges humans to keep up with the high evolutionary rate of bacteria. However, HGT is often unsuccessful because of barriers presented by requirements that must be fulfilled by the receiving organism, including the need for stable integration of the gene into the genome, no disturbance of other genetic structures or regulation, expression of the gene, and subsequent production of a functional protein, which requires correct folding and possibly modifications (32). In the present report we describe an HGT case study in which the transcription activator protein ComK from Bacillus subtilis was introduced into the heterologous host Lactococcus lactis. Previous research demonstrated that introduced ComK is produced in L. lactis and forms a functional protein, which can activate transcription of a ComK-dependent reporter fusion, albeit to a lower extent (31a). This implies that three of the requirements for successful HGT are fulfilled (stable integration, gene expression, and production of a functional protein). Thus, ComK in L. lactis serves as an excellent model to investigate the effects of introduction of a functional transcriptional regulator on the transcription profile of the host cell, which harbors multiple recognition sites for this protein. In order to fully understand the potential of ComK regulation in L. lactis, some background information is described first.
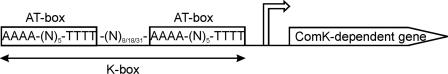
In the gram-positive soil bacterium B. subtilis, ComK is the key regulator of development of competence, a differentiation state that enables the cell to take up DNA from the environment and to incorporate this DNA into its genome. The competent state is characterized by drastic physiological changes in the cell, such as switching off DNA replication, cell wall synthesis, and cell division (13). Furthermore, a complex DNA binding, uptake, and integration machinery is synthesized, which makes the cell competent for transformation. Development of competence is tightly regulated; only when the free ComK level increases, at the onset of the stationary growth phase, can competence develop (for reviews see references 8 and 16). ComK activates gene transcription by binding to ComK-binding sites, so-called K-boxes, located upstream of ComK-regulated genes (14). Each K-box consists of two AT-boxes (consensus sequence, AAAA-[N]5-TTTT), which are separated by two, three, or four helical turns (Fig. 1). The genome of B. subtilis contains over 1,000 putative K-boxes, 30% of which are located in intergenic regions (15). These numbers reflect the large potential for ComK to affect transcriptional regulation.
FIG. 1.
Overview of a K-box. ComK-regulated genes are characterized by the presence of a K-box upstream of their promoter (arrow). Each K-box consists of two AT-boxes, which are separated by a flexible spacing. This results in positioning of both AT-boxes on the same side of the DNA helix with an interval of two, three, or four helical turns, depending on the size of the spacing, between the start of the two AT-boxes.
Interestingly, the genome of L. lactis contains around 1,350 putative K-boxes, roughly 30% of which (almost 400 K-boxes) are located in promoter regions. This number is considerably higher than the 474 K-boxes (on average) which would be expected to occur in random DNA with the length and A+T content of the L. lactis genome, suggesting that the K-boxes form a relevant feature on the genome. However, despite the presence of K-boxes, as well as homologues of some of the known competence genes, L. lactis does not contain comK itself, which brings into question the functionality of these K-boxes. Thus, introduction of B. subtilis ComK into L. lactis could result in extensive interference with transcriptional regulation of the host cell by direct ComK binding to a large number of K-boxes. The implications of this HGT event for the L. lactis transcription profile were investigated by DNA microarray studies, in which we compared a wild-type L. lactis strain and two ComK-producing strains, expressing either wild-type ComK (wtComK) or ComKΔC25, a transcription activation-deficient but DNA-binding mutant. Both ComK variants affected the transcription profiles of about 200 genes, and the majority were downregulated. The frequency of occurrence of a K-box within 200 bp from the start of a gene was higher for downregulated genes than for upregulated genes, suggesting that ComK has a repressive effect on gene transcription in L. lactis, mainly because of binding to available K-boxes. However, the majority of the regulated genes were indirectly affected as a consequence of ComK production in the cell. The results show that HGT does not necessarily also transfer the expected functionality of a gene and that the normal transcriptional regulation of the host cell can be severely affected.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains that were used in this study are listed in Table 1. L. lactis strains were grown at 30°C in 0.5× M17-based medium (Difco) supplemented with 0.5% glucose (GM17) and, if required, chloramphenicol and/or erythromycin (both at a concentration of 4 μg/ml). ComK expression was achieved by induction of the nisin-inducible promoter with 1:1,000 dilutions of supernatant of an overnight culture of the nisin-producing strain L. lactis NZ9700, which was grown in GM17 medium (7).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant feature(s) | Reference |
|---|---|---|
| Strains | ||
| L. lactis NZ9000 | MG1363 pepN::nisRK | 24 |
| L. lactis NZ9700 | Nisin producing, containing Tn5276 | 23 |
| Plasmids | ||
| pNZ8048 | Cmr, inducible expression vector containing the nisA promoter | 24 |
| pNZ-His6-ComK | Cmr, his6-comK under control of PnisA | 31a |
| pNZ-ComKΔC25 | Cmr, his6-comKΔC25 under control of PnisA | 31a |
| pG-wt | Eryr, contains wild-type common K-box of comG promoter | 31a |
| pORI::PctrA | Eryr; pORI13 carrying a 233-bp ctrA promoter fragment | 6 |
| pPywcC | Eryr, contains PywcC-lacZ fusion | This study |
| pPtig | Eryr, contains Ptig-lacZ fusion | This study |
| pPctrA | Eryr, contains PctrA-lacZ fusion | This study |
DNA manipulations, materials, and transformations.
Standard molecular biological techniques were performed as described previously (1, 30). Enzymes were purchased from Roche, New England Biolabs, or Pharmacia. Plasmids were isolated using a High Pure plasmid isolation kit (Roche), and PCR products were purified with a High Pure PCR product purification kit (Roche). L. lactis strains were transformed by electrotransformation using a gene pulser (Bio-Rad Laboratories) as described by Leenhouts and Venema (27).
PCR amplification and plasmid construction.
PCRs were performed as described by Innes and Gelfand (20), using Pwo or Expand DNA polymerase (Roche) with chromosomal DNA of B. subtilis 168 or L. lactis MG1363 as the template. Plasmids used in this study are listed in Table 1. To confirm the effects on transcription of L. lactis genes demonstrated by DNA microarray analyses, around 200 bp of the promoter region of two model genes was fused to a promoterless lacZ gene in plasmid pILORI4. For the ywcC gene, the promoter region was amplified using primers ywcC-start (5′-GATCGAATTCGAAAGCTATCCTACCCCCCTTTC) and ywcC-end (5′-GATCTCTAGATTAAGATACACGTTTAGTATAACCGCC). The tig promoter region was amplified using primers tig-start (GATCGAATTCTATGACTAAGCTAAGCCCTGG) and tig-end (GATCTCTAGATTAGAGTGTACCCTTAGTATCACTAG). In all primers, the annealing sequence is underlined. The resulting PCR products were digested with XbaI and EcoRI and ligated into XbaI/EcoRI-digested pILORI4, yielding plasmids pPywcC and pPtig, respectively. As a control for the cutoff level of the DNA microarray data, a ctrA-lacZ fusion was tested. A 242-bp fragment comprising the upstream region of ctrA was cut from plasmid pORI::PctrA (6) using XbaI and EcoRI, and the fragment was introduced into pILORI4, resulting in plasmid pPctrA.
ComK production and transcription activation assays.
B. subtilis ComK was produced in L. lactis NZ9000, an MG1363 derivative, using plasmid pNZ-His6-ComK for wild-type ComK or plasmid pNZ-His6-ComKΔC25 for a C-terminal truncation variant of ComK. These plasmids contained the comK gene under control of the nisin-inducible promoter, and induction with nisin resulted in the synthesis of wtComK and ComKΔC25, respectively. Both ComK proteins were synthesized with an N-terminal His tag.
To determine the activity of the ComK proteins produced, transcription activation assays were performed using the comG-promoter-lacZ fusion on plasmid pG-wt as a reporter. For this purpose, ComK expression was induced by adding nisin-containing supernatant after 3 h of growth. Samples for β-galactosidase assays were taken from the moment of induction until 2 h after induction at 30-min intervals. β-Galactosidase activity was determined as described by Israelsen et al. (21). The same experiments were performed using the promoter-lacZ fusion on plasmids pPywcC, pPtig, and pPctrA as the reporters. ComK production levels were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (26) and Western blot analysis (33), using a His tag-specific first antibody and an anti-mouse horseradish peroxidase secondary antibody (both obtained from Amersham). The signals were visualized by chemiluminescent detection using the ECL Western blotting analysis system from Amersham.
RNA isolation, cDNA labeling, and hybridization.
To determine the effect of wtComK production on the transcription profile of L. lactis, cultures of L. lactis NZ9000 with plasmid pNZ8048 and with plasmid pNZ-His6-ComK were grown for 3 h until the end of the exponential growth phase and were then induced with a 1:1,000 dilution of a supernatant of the nisin-producing strain L. lactis NZ9700. After 2 h of induction, cells from 25 ml of the culture were harvested by centrifugation (1 min, 8,000× g, Eppendorf centrifuge) and frozen in liquid nitrogen. Three biological replicates were performed under identical conditions. mRNA was isolated from the cells of each culture as described previously (35). cDNA was obtained by reverse transcription and labeled with Cy3 or Cy5. As a control, dye swap reactions were performed under the same conditions. The labeled cDNA samples were hybridized onto an L. lactis MG1363 microarray slide as described previously (25, 35). To determine the effect of ComKΔC25 on the transcription profile of L. lactis, the procedure described above was repeated with cultures of L. lactis NZ9000 with plasmid pNZ8048 and with plasmid pNZ-His6-ComKΔC25.
Bioinformatic analyses.
After the signals on the slides were scanned, bad spots and spots whose intensities were too low were removed from the data sets using Array Pro analyzer 4.5 (MediaCybernetics, Gleichen, Germany). Based on the assumption that expression of most of the genes is not different in the two situations compared, the Cy3/Cy5 ratios were normalized using a grid-based Lowess fit (37). Further analysis was performed using the MicroPrep software package, with a subsequent PrePrep, Prep, and PostPrep analysis (10, 34) and a t test (17). The MicroPrep software package can be requested at http://molgen.biol.rug.nl/molgen/research/molgen software.php.
Identification of functional categories among regulated genes.
Functional categories of the regulated genes were identified using the Functional Information Viewer and Analyzer (FIVA) program (E. J. Blom, D. W. J. Bosman, S. A. F. T. van Hijum, L. Tijsma, J. B. T. M. Roerdink, and O. P. Kuipers, submitted for publication).
Genomic overview and K-box positioning.
The Genome2D software package (2) was used to create an overview of the positions of regulated genes on the genome of L. lactis MG1363. Furthermore, the program was used to determine the position of the nearest K-box for each gene.
RESULTS
B. subtilis ComK is active in L. lactis and affects culture growth.
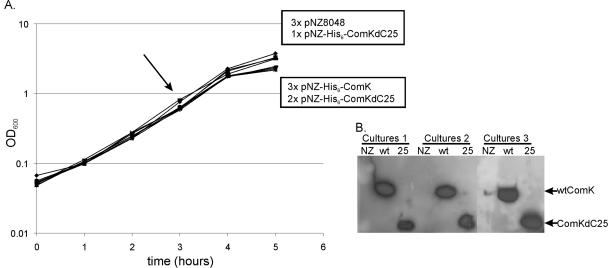
The competence transcription factor ComK activates gene transcription in B. subtilis by binding to K-boxes in the upstream region of ComK-dependent genes. Like the genome of B. subtilis, the genome of L. lactis contains a large number of K-boxes, which are potential targets for transcription regulation by ComK. However, L. lactis lacks a comK gene itself. Previous research demonstrated the activity of B. subtilis ComK in L. lactis by showing transcription activation at a B. subtilis comG-promoter-lacZ fusion (31a). In principle, ComK of B. subtilis could also regulate transcription from the available K-boxes when it is expressed in L. lactis. To test this hypothesis, DNA microarray analyses were performed by comparing the transcription profiles of L. lactis NZ9000 with plasmid pNZ8048 and with plasmid pNZ-His6-ComK, both grown in the presence of nisin-containing supernatant to induce transcription from the nisin-inducible promoter on the plasmids. In order to obtain significant array data, three biological replicate samples of each strain were produced. Until the moment of induction, after 3 h of growth, all cultures displayed similar growth curves. However, upon induction of ComK production the growth rates of the three cultures of L. lactis NZ9000(pNZ-His6-ComK) were reduced, indicating that comK expression affects growth of the host cells (Fig. 2A). Western blots demonstrated that ComK was produced at similar levels, as shown previously (Fig. 2B) (31a).
FIG. 2.
Growth and ComK production of the strains tested. (A) Independent cultures of L. lactis NZ9000(pNZ8048), L. lactis NZ9000(pNZ-His6-ComK), and L. lactis NZ9000(pNZ-His6-ComKΔC25) were grown as biological replicates. The moment of induction with supernatant of nisin-producing strain L. lactis NZ9700 at a 1:1,000 dilution is indicated by an arrow. Two growth rates were distinguished after induction of ComK production. For the organisms in the upper box the growth rates of the cultures were normal; for the organisms in the lower box the growth rates of the cultures were decreased. Samples for RNA isolation were harvested after 2 h of induction. OD600, optical density at 600 nm. (B) Western blotting was performed to detect the His-tagged ComK proteins using an anti-His antibody. Samples of three cultures per strain were loaded on a gel. Lanes NZ, L. lactis NZ9000(pNZ8048); lanes wt, L. lactis NZ9000(pNZ-His6-ComK); lanes 25, L. lactis NZ9000(pNZ-His6-ComKΔC25).
ComK production affects 7% of the L. lactis genome.
To determine the effect of the introduction of B. subtilis ComK on the transcription profile of the L. lactis host cell, a transcriptome analysis was performed. Including dye swaps for each of the three biological replicates, six slides were used, and wild-type L. lactis and a ComK-producing strain were compared. The transcription profile of the latter strain revealed significant effects on about 200 genes, corresponding to 7% of the genome. Notably, 89 genes (3.0% of the genome) were upregulated, while 114 genes (4.0% of the genome) were expressed less in the presence of ComK. The up- and downregulated genes with the greatest differences are listed in Tables 2 and 3, respectively.
TABLE 2.
Top 30 upregulated genes for wtComK or ComKΔC25 expressiona
| Gene | Description | Ratio of transcription
|
K-box | |
|---|---|---|---|---|
| wtComK | ComKΔC25 | |||
| dacA | d-Alanyl-d-alanine carboxypeptidase | 9.8 | 5.5 | |
| orf1837 | Hypothetical protein | 8.8 | 11.2 | |
| orf258 | Hypothetical protein | 8.2 | 13.0 | |
| yriC | Hypothetical protein | 6.5 | 15.9 | Yes |
| ydaG | ABC transporter, ATP-binding and permease protein | 6.3 | 5.7 | |
| dnaG | DNA primase | 6.2 | 3.9 | Yes |
| yjaE | Hypothetical protein | 6.2 | 5.4 | |
| guaC | GMP reductase | 5.0 | 2.4 | Yes |
| cadA | Cadmium efflux ATPase | 4.9 | 3.7 | |
| yrbB | Hypothetical protein | 4.7 | 2.2 | |
| yhfA | Hypothetical protein | 4.7 | 2.6 | |
| ymfD | Integrase-recombinase | 4.7 | 4.2 | |
| yriD | Hypothetical protein | 4.6 | 4.7 | Yes |
| adhE | Alcohol-acetaldehyde dehydrogenase | 4.5 | 6.3 | |
| ydbA | ABC transporter, ATP-binding and permease protein | 4.3 | 4.7 | |
| yleF | Transcription regulator | 4.1 | 2.2 | |
| ycgD | Oxidoreductase | 4.0 | 3.5 | |
| vacB1 | RNase | 4.0 | 3.5 | |
| trxH | Thioredoxin, H-type | 3.8 | 2.0 | |
| orf1734 | Hypothetical protein | 3.7 | ||
| ytjD | Hypothetical protein | 3.7 | 6.0 | |
| yuiE | Hypothetical protein | 3.6 | ||
| busAA | OpuAA | 3.4 | 3.5 | Yes |
| ywjF | 3-Hydroxyisobutyrate dehydrogenase | 3.4 | ||
| purM | Phosphoribosylformylglycinamide cycloligase | 3.2 | ||
| yccL | Hypothetical protein | 3.1 | ||
| ypaD | Hypothetical protein | 3.1 | 6.6 | |
| yejC | Hypothetical protein | 3.1 | ||
| yxdB | Hypothetical protein | 3.1 | 2.4 | Yes |
| ycdG | Hypothetical protein | 3.1 | 16.0 | |
The list of genes is based on the results for a wtComK-producing strain. For each gene, the putative function and the ratio of transcription relative to L. lactis NZ9000(pNZ8048) is indicated for wtComK and, if regulated, for ComKΔC25. The presence of a K-box within 200 bp from the transcription start site is also indicated.
TABLE 3.
Top 30 downregulated genes for wtComK or ComKΔC25 expressiona
| Gene | Description | Ratio of transcription
|
K-box | |
|---|---|---|---|---|
| wtComK | ComKΔC25 | |||
| agl | Alpha-glucosidase | −15.1 | −22.2 | |
| mapA | Maltose phosphorylase | −13.4 | −7.8 | |
| ywcC | Hypothetical protein | −11.4 | −5.1 | Yes |
| pepC | Aminopeptidase C | −11.1 | ||
| dexC | Neopullulanase | −11.0 | −7.7 | |
| malE | Hypothetical protein | −7.9 | −7.0 | |
| tig | Trigger factor | −7.7 | −2.0 | Yes |
| amyY | Alpha-amylase | −6.1 | −7.5 | |
| ytaA | Hypothetical protein | −5.4 | −15.6 | |
| ydbF | Transcription regulator | −5.2 | −7.5 | |
| yjG | Hypothetical protein | −5.2 | −3.5 | Yes |
| cydA | Cytochrome d ubiquinol oxidase subunit I | −5.1 | −2.3 | |
| pgmB | Metabolic protein | −4.6 | −5.5 | Yes |
| ydbH | Hypothetical protein | −4.5 | −8.6 | |
| dnaN | DNA polymerase III, beta chain | −4.5 | −2.0 | |
| yjaB | Oxidoreductase | −4.2 | −3.2 | |
| orf1188 | Hypothetical protein | −4.2 | −4.1 | |
| osmC | Osmotically induced protein | −4.2 | −5.0 | |
| maa | Maltose O-acetyltransferase | −4.1 | −4.2 | |
| yvdD | Hypothetical protein | −4.0 | −5.2 | |
| ffh | Signal recognition particle protein | −4.0 | −3.3 | |
| nusB | Transcription termination protein | −4.0 | ||
| pepN | Aminopeptidase N | −3.8 | −4.8 | |
| malG | Hypothetical protein | −3.8 | −3.0 | |
| dexA | Oligo-1,6-glucosidase | −3.8 | −3.8 | |
| amtB | NgrA-like protein | −3.7 | −2.9 | |
| yseI | Hypothetical protein | −3.6 | −3.0 | |
| yjbD | Hypothetical protein | −3.6 | −3.0 | |
| pdhD | Lipoamide dehydrogenase component of PDH complex | −3.5 | ||
| accA | Acetyl conenzyme A carboxylase carboxyl transferase α-subunit | −3.5 | −2.6 | |
See Table 2, footnote a.
Genomic organization of ComK-affected genes.
The genomic organization of the ComK-affected genes was visualized using the Genome2D software package (2). Production of ComK was shown to influence transcription of mainly individual or paired genes, but some larger operons could also be discerned. The most eye-catching operon consists of dexCA, maa, amyY, agl, and mapA, which are among the top downregulated genes. The gene products are probably involved in sugar metabolism of the cell. Downregulation of this operon can therefore probably be explained by the reduced growth rate of the ComK-producing strain upon induction of ComK expression (Fig. 2A). Furthermore, some members of the opp operon are downregulated probably also because of the effects of ComK expression on the growth rate of L. lactis. Other clearly downregulated operons are the ycgFGHIJ operon, the rnc-smc-yibCD operon, and the ndrHIE operon. Only two relatively small upregulated operons were observed, the ydaFG-ydbA operon and the ydjB-trxH-ydjD operon.
Most of the ComK effects on the transcription profile are indirect.
An interesting question concerning the effects of production of ComK on the transcription profile is whether these effects are direct or indirect. Direct effects are considered to be caused by binding of ComK to a ComK-binding site, i.e., a K-box, upstream of a gene. Previous research with B. subtilis on the predictive value of the presence of a K-box for regulation by ComK revealed that K-boxes that had 13 or more matches with the 16-bp consensus sequence and were located within 200 bp upstream of the start of a gene could be good targets for regulation by ComK (15). In the present study, a list of K-boxes upstream of the ComK-affected genes was generated, taking into account the important characteristics determined in B. subtilis (Tables 2 and 3). When this list was considered, we concluded that the majority of the effects of ComK expression are indirect, since only 11 of the 89 upregulated genes (12.3%) and 31 of the 114 downregulated genes (27.2%) contain a K-box. Transcriptome analyses of B. subtilis indicated that the frequency of occurrence of a K-box is higher for ComK-activated genes and operons (45%) (15), while no significant repression by ComK has been reported (3, 15, 28).
The low percentage of genes directly affected by ComK raises questions about the nature of the ComK-induced effects. In order to obtain more information about the background of these effects, functional categories of regulated genes were searched by using the software package FIVA. An overview is shown in Table 4. The FIVA analysis demonstrated that the upregulated genes were involved mainly in protein synthesis. The downregulated genes are grouped into two main categories, ATP binding and transport, and three smaller groups involved in carbohydrate metabolism, regulation of transcription, and signal transduction. The categories observed suggest that the induced effects result mainly from the reduced growth in a ComK-producing culture.
TABLE 4.
Organization of regulated genes in functional categoriesa
| Functional category | No. of genes (no. of genes with K-box)
|
|
|---|---|---|
| Downregulated | Upregulated | |
| ATP binding | 17 (5) | 5 (1) |
| Membrane/transport | 20 (3) | 11 (1) |
| Carbohydrate metabolism | 7 (0) | 3 (0) |
| Regulation of transcription | 7 (2) | 2 (0) |
| Signal transduction | 6 (2) | 0 (0) |
| Protein biosynthesis | 1 (1) | 5 (0) |
Using the FIVA program, regulated genes were grouped into functional categories. For each functional category the number of genes that are downregulated or upregulated in the presence of wtComK is indicated, as is the number of the genes that contain a K-box.
ComKΔC25 affects transcription less than wtComK.
The most intriguing observation concerns the fact that most of the direct effects exerted by ComK involve downregulation of gene expression, although ComK in B. subtilis acts almost exclusively as a transcription activator (3, 15, 28). The extensive downregulation of gene transcription in L. lactis might result from binding of ComK to available K-boxes. Following this reasoning, ComK would disturb the normal transcription regulation by binding to K-boxes. To test this hypothesis, a similar DNA microarray experiment was performed with a transcription activation-deficient ComK variant, His6-ComKΔC25. Previous research demonstrated that transcription activation by this mutant is completely abolished, while DNA binding is retained with wild-type affinity, despite binding as dimers instead of tetramers (31a). The level of expression of ComKΔC25 was checked on Western blots (Fig. 2B), and the results demonstrated that the protein level was similar to that for wtComK. Transcriptome analyses demonstrated that production of ComKΔC25 and production of wtComK affected the transcription profile of L. lactis tosimilar extents. Altogether, 237 genes (8.3% of the genome) were affected in the presence of ComKΔC25; this included 105 upregulated genes (3.7% of the genome) and 132 downregulated genes (4.6% of the genome).
Comparison of the genes affected by wtComK and ComKΔC25 demonstrated that the majority of the top 30 up- and downregulated genes were regulated in both ComK-producing strains (Tables 2 and 3). Overall, 56.6% of the wtComK-affected genes were also affected by ComKΔC25. Given the high levels of similarity of the top regulated genes, this percentage seems relatively low. However, the fact that the differences occur mainly in the genes that are regulated less, which might just miss the cutoff for fold regulation or significance in either of the two lists, should be considered. Furthermore, without exception, genes upregulated in a wtComK expression strain are also upregulated or unchanged by ComKΔC25 production and vice versa, but they are never downregulated. The same is true for the downregulated genes. The great similarity of the transcription profiles of wtComK and ComKΔC25 suggests that ComK influences gene regulation by binding to available K-boxes, which interferes with normal gene transcription, thereby explaining the direct down effects. More often, however, ComK production, which probably causes various stress responses, results in a large number of indirect effects.
β-Galactosidase assays show repression by both ComK variants.
To confirm the negative effect of ComK on transcription of L. lactis genes, promoter-lacZ fusions were made for three target genes, ywcC, tig, and ctrA. The ywcC and tig genes were chosen because they are the two strongest ComK-repressed genes with a K-box within 200 bp upstream of the starting position. Transcription of ywcC and tig was affected by both wild-type and mutant ComK. The ctrA gene, on the other hand, was chosen because it is a gene that is only mildly affected by ComK, as determined by the DNA microarray analysis (−1.7-fold by wtComK and −1.2-fold by ComKΔC25). It can thus serve as a proper control to determine whether the effects of ComK on genes that are regulated at a low level, as observed in the DNA microarrays, are really significant. A β-galactosidase assay, using plasmid pPywcC as a reporter, demonstrated that the level of transcription from the ywcC promoter is high in the absence of ComK and is reduced when wtComK is produced (Fig. 3A). A similar reduction in the transcription level is seen when ComKΔC25 is expressed. Also, in the case of pPtig, production of ComK reduces the transcription of the reporter gene, although in this case the effect is clearly stronger for wtComK than for ComKΔC25 (Fig. 3B). A ctrA-lacZ reporter revealed mild effects of both ComK variants on ctrA transcription (Fig. 3C), confirming the small difference that was below the cutoff observed in the DNA microarray study. This result implies that the genes regulated at a low level, as observed in the DNA microarrays, do reflect effects of ComK on gene transcription regulation, thereby demonstrating the significance of these data. Although the ratio of transcription of ywcC in wild-type L. lactis to transcription of ywcC in the ComK expression strains is less than the ratio indicated in Table 3, these results confirm the trend that was observed when DNA microarray analyses were performed.
FIG. 3.
ComK negatively affects transcription of ywcC and tig. β-Galactosidase (Betagal) assays were performed to verify the negative effects of ComK production on transcription of ywcC and tig, using a PywcC-lacZ fusion (A) and a Ptig-lacZ fusion (B) as reporters, respectively. As a control for significant regulation, the mild effect of ComK on PctrA-lacZ, which just missed the arbitrary cutoff level, was determined (C). At the end of the exponential growth phase, expression of wtComK or ComKΔC25 was induced. β-Galactosidase activity was determined at 30-min intervals until 2 h after induction. For each strain, four independent cultures were grown, and the average activity (in Miller units) is shown along with the standard deviation for the average. Symbols: ⧫, no ComK (empty plasmid pNZ8048); ▴, wtComK; ▪, ComKΔC25.
DISCUSSION
Bacteria undergo continuous evolution of their genotypes. A possible mechanism for genotypic changes is HGT, which has been reported for a wide variety of bacteria (31, 32). In this paper we describe a case study of HGT by introduction of the competence transcription factor ComK of B. subtilis into the recipient L. lactis. A successful HGT event should meet several criteria, including proper expression of the introduced gene and subsequent production of a functional protein (32). Previous research demonstrated that our model fulfils these criteria, since B. subtilis ComK that is produced in L. lactis can activate transcription of a ComK-dependent gene in the same cell, albeit at a low level (31a). Interestingly, L. lactis lacks a comK homologue but does contain a large number of K-boxes and homologues of some of the late competence genes, reflecting a large potential for the introduced B. subtilis ComK to act on the host's transcription profile. DNA microarray analyses were performed to investigate these effects, and the results demonstrated that production of ComK affects transcription of about 200 genes in L. lactis (89 upregulated genes and 114 downregulated genes). Only about 12% of the upregulated genes and 27% of the downregulated genes contain a K-box in the upstream DNA region, indicating that the majority of ComK-induced effects are indirect. The FIVA software package was used to group the affected genes into functional categories, confirming that regulation results mainly from the effects of ComK production on the cell. For example, ComK expression causes downregulation of genes involved in carbohydrate metabolism, which can be explained by the reduced growth of ComK-producing cultures. Another important category consists of genes involved in protein synthesis, which are clearly upregulated in a ComK-expressing strain. One possible explanation for this upregulation is provided by the direct downregulation of other genes by ComK. The products of the latter genes are depleted from the cell, which might react by enhancing protein synthesis in order to still produce the required proteins. When the lists of regulated genes in L. lactis are compared with the lists resulting from a comparable analysis for B. subtilis, there is hardly any overlap in regulated genes, although in B. subtilis genes involved in metabolic pathways and stress responses are activated upon ComK synthesis. However, the majority of ComK-activated genes in B. subtilis are related to competence development, while in L. lactis no homologues of known competence genes are affected by ComK production, indicating that HGT of comK does not necessarily also imply transfer of activation of competence genes.
Although most ComK effects are indirect, some direct effects could be discerned as well. The most striking observation is that direct transcription regulation mainly involves downregulation of gene transcription, which was concluded from the fact that the frequency of occurrence of a K-box was much higher for downregulated genes (27%) than for upregulated genes (12%). This is a remarkable difference from the situation in B. subtilis, since in this species no significant downregulation of gene transcription by ComK has been reported (3, 15, 28). As an explanation, interference with gene transcription due to binding of ComK to available K-boxes is assumed. This hypothesis was confirmed by an additional DNA microarray study using a transcription activation-deficient but DNA-binding ComK variant, ComKΔC25. The extensive overlap in the lists of genes affected in the two ComK-producing strains confirmed that the direct effects of ComK on the transcription profile of L. lactis are mainly caused by selective positional binding of ComK, thereby disturbing normal transcription activation. The negative effects of both wild-type and mutant ComK on transcription of L. lactis genes were further demonstrated using β-galactosidase assays with promoter-lacZ fusions of ywcC and tig, the two L. lactis genes preceded by a K-box affected the most, as reporters (Fig. 3A and B). These experiments confirmed that there was transcriptional repression upon ComK production, most likely via direct binding to the K-box, thereby obstructing transcription activation. A similar study using a PctrA-lacZ fusion showed that even the small ComK effect on ctrA transcription was reflected by the results of β-galactosidase assays (Fig. 3C), confirming the significance of the observed ComK effects on genes affected at a low level displaying values just below the arbitrary cutoff levels of twofold up- or downregulation in DNA microarray studies (Tables 2 and 3).
The similarity in upregulated genes between the expression of wild-type ComK and the expression of mutant ComK suggests that the effects are indirect rather than direct also for the 12% of the genes that do contain a K-box. The occurrence of a K-box upstream of these genes is therefore expected to be a coincidence instead of a biologically relevant feature of the regulated genes. It might be concluded from the results described here that ComK has only a repressing effect on gene transcription in L. lactis. However, despite the fact that direct activation was not observed in the transcription profile of L. lactis, this possibility cannot be excluded. Previous research has demonstrated that there was transcription activation by wtComK at the comG promoter of B. subtilis when it was introduced into L. lactis, but the level of transcription was at least 10 times lower than that at the same promoter in B. subtilis, suggesting that the conditions in L. lactis do not allow optimal transcription activation by ComK, which could, for example, be due to differences in RNA polymerase. Although speculative, it is likely that ComK of B. subtilis does activate transcription by binding to K-boxes in L. lactis but that the transcription level in many cases is too low to be discerned by transcriptome analyses among the large indirect effects induced by production of ComK.
Returning to the discussion of HGT, the most intriguing observation in this study is undoubtedly the change of ComK from a clear activator in B. subtilis into a mainly repressing protein in L. lactis. This observation demonstrates that a known function of a protein cannot automatically be extrapolated to other potential targets for this protein in another organism. It has been reported previously that observed HGT events involve operational genes (e.g., housekeeping genes) more often than they involve informational genes (e.g., genes involved in transcription and translation processes) (29). An explanation for this is provided by the complexity hypothesis, which states that the more complex the role of the introduced gene, the smaller the chance for successful HGT (22). Informational genes often are parts of larger structures, implying that for proper functioning not just one gene but a set of genes should be transferred. This may also play a role in the functionality of regulator genes in their new background, since to be functional, they require the presence of target sequences and relevant genes under control of these sequences. The previously described examples of successful HGT of regulator genes most often involved transfer between very closely related species and/or transfer of complete pathogenicity islands or plasmids, including a regulator and its target genes. An intriguing example is the Bacillus cereus group of organisms, including B. anthracis, B. cereus, and Bacillus thuringiensis. Based on chromosome sequences, these species are basically indistinguishable and should actually be considered one species (18). Their functional differences are located mainly on large plasmids, for which horizontal spreading has been shown among members of the B. cereus group, resulting in phenotypically changing a B. cereus strain into B. thuringiensis and even B. anthracis-like strains (12, 19). This can have serious consequences for humans considering the pathogenicity of these species.
Based on the observation that successful HGT of regulator genes often involves the transfer of complete plasmids, it might not be surprising that in our model, in which only the regulatory gene was introduced, comK is not functional in L. lactis, as it is in B. subtilis. Not only does the introduced gene apparently lack its original functionality, but in addition, the expression of a potentially pleiotropic regulator might induce extensive stress responses and growth defects in the host organism, thereby interfering with the normal behavior of the cell. This study presents a novel way of looking at an HGT event by thoroughly investigating the effects of introduction of a foreign gene on the transcription profile of the recipient bacterium. Using this approach, we demonstrated that HGT of a regulatory gene can be an event with highly counterintuitive and unexpected effects compared to the original function of the gene.
Acknowledgments
We thank Naomi Kramer for help with performing the DNA microarray analyses and analyzing the data obtained. Furthermore, we thank Andrzej Lulko, Aldert Zomer, and Sacha van Hijum for help with the data analysis and Evert-Jan Blom and Anne de Jong for help with the identification of functional categories among the regulated genes.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidham, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 2.Baerends, R. J. S., W. K. Smits, A. de Jong, L. W. Hamoen, J. Kok, and O. P. Kuipers. 2004. Genome2D: a visualization tool for rapid analysis of bacterial transcriptome data. Genome Biol. 5:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin, A., B. Quinquis, A. Sorokin, and D. S. Ehrlich. 2005. Recent genetic transfer between Lactococcus lactis and enterobacteria. J. Bacteriol. 186:6671-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson, J. 1999. Genetic exchange between bacteria and environment. Plasmids 42:73-91. [DOI] [PubMed] [Google Scholar]
- 6.Den Hengst, C. D., S. A. F. T. van Hijum, J. M. W. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. [DOI] [PubMed] [Google Scholar]
- 7.De Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau, D., and C. M. J. Lovett. 2002. Transformation and recombination, p. 453-471. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed), Bacillus subtilis and its closest relatives. From genes to cells. American Society for Microbiology, Washington, D.C.
- 9.Dufraigne, C., B. Fertil, S. Lespinats, A. Giron, and P. Deschavanne. 2005. Detection and characterization of horizontal transfers in prokaryotes using genomic signature. Nucleic Acids Res. 33:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia de la Nava, J., S. van Hijum, and O. Trelles. 2003. Prep: gene expression data pre-processing. Bioinformatics 19:2328-2329. [DOI] [PubMed] [Google Scholar]
- 11.Gill, S. R., D. E. Fouts, G. L. Archer, et al. 2005. Insight on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epodermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzales, J. M. J., B. S. Brown, and B. C. Carlton. 1982. Transfer of Bacillus thuringiensis plasmid coding for δ-endotoxin among strains of Bacillus thuringiensis and Bacillus cereus. Proc. Natl. Acad. Sci. USA. 79:6951-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haijema, B. J., J. Hahn, J. Haynes, and D. Dubnau. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40:52-64. [DOI] [PubMed] [Google Scholar]
- 14.Hamoen, L. W., A. F. van Werkhoven, J. J. E. Bijlsma, D. Dubnau, and G. Venema. 1998. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 12:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9-17. [DOI] [PubMed] [Google Scholar]
- 17.Hatfield, G. W., S. Hung, and P. Baldi. 2003. Differential analysis of DNA microarray expression data. Mol. Microbiol. 47:871-877. [DOI] [PubMed] [Google Scholar]
- 18.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmaster, A. R., J. Ravel., D. A. Rasko, et al. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101, 8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Innes, M. A., and D. H. Gelfand. 1990. Optimization of PCRs, p. 3-12. In M. A. Innes, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 21.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Evolution 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 24.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum-sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 25.Kuipers, O. P., A. de Jong, R. J. S. Baerends, S. A. F. T. van Hijum, A. L. Zomer, H. A. Karsens, C. D. den Hengst, N. E. Kramer, G. Buist, and J. Kok. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Leeuwenhoek 82:113-122. [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Leenhouts, K. J., and G. Venema. 1993. Plasmids, a practical approach. Oxford University Press, Oxford, United Kingdom.
- 28.Ogura, M., H. Yamaguchi, K. Kobayashi, N. Ogasawara, Y. Fujita, and T. Tanaka. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivera, M. C., R. Jain, J. E. Moore, and J. A. Lake. 1998. Genomic evidence for two functionally distinct gene classes. Genetics 95:6239-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sørensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal gene transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700-710. [DOI] [PubMed] [Google Scholar]
- 31a.Susanna, K. A., F. Fusetti, A.-M. W. H. Thunnissen, L. W. Hamoen, and O. P. Kuipers. The C-terminal region of the competence transcription factor ComK of Bacillus subtilis is required for transcription activation. Microbiology, in press. [DOI] [PubMed]
- 32.Thomas, C. M., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 33.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Hijum, S. A. F. T., J. Garcia de la Nava, O. Trelles, J. Kok, and O. P. Kuipers. 2003. MicroPrep: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2:241-244. [PubMed] [Google Scholar]
- 35.Van Hijum, S. A. F. T., R. J. Baerends, H. A. Karsens, A. de Jong, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. A generally applicable validation scheme for the assessment of factors involved in reproducibility of DNA microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed]
- 36.Wang, Y., G. R. Wang, N. B. Shoemaker, T. R. Whitehead, and A. A. Salyers. 2005. Distribution of the ermG gene among bacterial isolates from porcine intestinal contents. Appl. Environ. Microbiol. 71:4930-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Workman, C., L. J. Jensen, H. Jarmer, R. Berka, L. Gautier, H. B. Nielsen, H. H. Saxild, C. Nielsen, S. Brunak, and S. Knudsen. 2002. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3:research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]