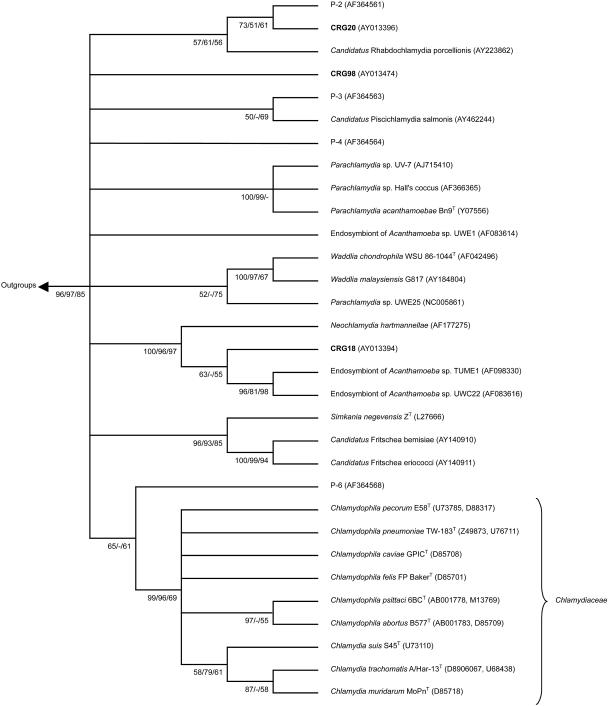
FIG. 1.
Phylogenetic relationships among Chlamydiales. The tree was inferred by using maximum-likelihood (ML), neighbor joining (NJ), and maximum-parsimony (MP) analysis of a 210- to 223-bp region of the 16S rRNA Chlamydiales signature sequence. The unrooted consensus tree topology is shown. Numbers indicate the percentage of times each branch appeared in a tree during 1,000 bootstrap samples (NJ and MP) or 10,000 puzzling steps (ML) in the order (NJ/MP/ML). Multifurcations connect branches for which the relative order could not unambiguously be determined. Branches supported by a bootstrap or puzzling reliability value of ≥50% with at least two treeing methods are shown. The GenBank accession numbers are given in parentheses. Included in the inference of the phylogenetic tree were (i) sequences of all type strains of classified Chlamydiales (T), (ii) all sequences of cultured chlamydia-like organisms (endosymbionts of Acanthamoeba sp. strains UWE1, TUME1, and UWC22, Parachlamydia sp. strain UWE25, Parachlamydia sp. strain UV-7, and Parachlamydia sp. strain Hall's coccus; endosymbionts of Hartmanella vermiformis and Neochlamydia hartmannellae; “Candidatus Rhabdochlamydia porcellionis”; Waddlia malaysiensis G817; “Candidatus Fritschea bemisiae”; and “Candidatus Fritschea eriococci”), (iii) cloned sequences from activated sludge that are representatives of four new environmental chlamydial lineages suggested by Horn and Wagner (15) (P-2, P-3, P-4, and P-6), (iv) cloned sequence of “Candidatus Piscichlamydia salmonis,” and (v) outgroup species Escherichia coli (GenBank accession number AE000460) and Rickettsia prowazekii (GenBank accession number M21789).