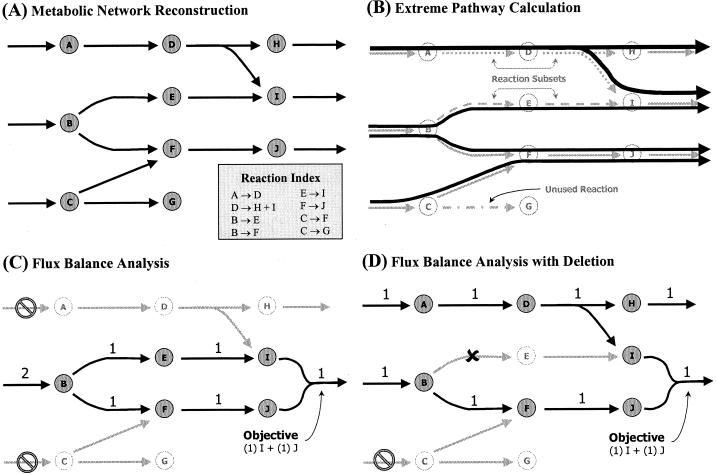
FIG. 1.
Overview of the in silico modeling methods and their conceptual basis. (A) Hypothetical network describing a set of metabolic reactions. Based on genomic, biochemical, and physiological data, such a reaction network can be reconstructed to represent the set of chemical reactions predicted to occur within an organism. (B) Extreme pathways are calculated for the reconstructed reaction network. Reactions that do not occur in any of the extreme pathways constitute unused reactions. Certain reactions always participate together when active in an extreme pathway. These groups of concomitantly occurring reactions constitute reaction subsets, as illustrated. (C) FBA is used to determine what input substrates and balanced reaction fluxes are required to meet the demand of producing metabolites I and J simultaneously in a fixed ratio. In this example only substrate B is available. (D) The reaction from metabolite B to E is eliminated, requiring the use of substrate A to meet the production demands of I and J. This situation also leads to the production of metabolite H as a by-product. Note that this reaction is essential if substrate A is not present.