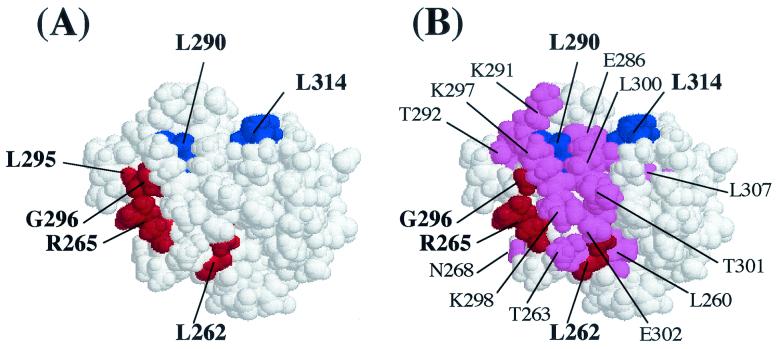
FIG. 5.
Space-filling model of the αCTD based on the atomic coordinates of Jeon et al. (24) showing the positions of some of the residues identified as important for LuxR-dependent (A) and LuxRΔN-dependent (B) activation of the lux operon. (A) Amino acid residues producing >2-fold effects on LuxR-dependent luciferase levels in vivo when changed to alanine are highlighted. The residues in red (262, 265, 295, and 296) are hypothesized to be involved in DNA recognition, and the residues in blue (290 and 314) are hypothesized to be involved in protein-protein interactions. (B) Amino acid residues producing >2-fold effect on either LuxRΔN-dependent luciferase levels in vivo or transcription rates in vitro are highlighted. The residues in red (262, 265 and 296) and blue (290 and 314) were found to be important in both LuxR- and LuxRΔN-dependent activation, while the residues in violet are unique to the LuxRΔN-dependent mechanism.