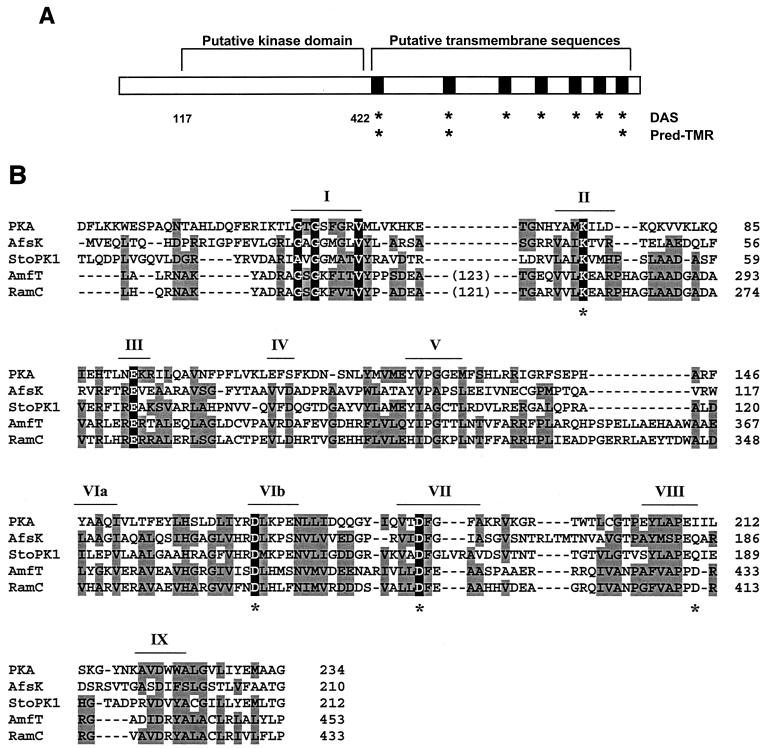
FIG. 1.
(A) Sequence analysis of the RamC polypeptide. Locations of the membrane-spanning sequences predicted by the DAS and Pred-TMR programs are indicated by asterisks. The putative kinase is bounded by residues 117 and 422. (B) Sequence alignment of RamC with a cAMP-dependent protein kinase (PKA), the S. coelicolor kinase AfsK, the Streptomyces toyocaensis kinase StoPK1, and the S. griseus RamC homologue AmfT. Residues shaded in grey denote similarity to RamC; those shaded in black are predicted, on the basis of well-known model kinases, to be involved in catalysis of phosphorylation. Conserved kinase-like sequence motifs (“Hanks” motifs) are indicated by roman numerals according to convention (3). Asterisks indicate the point mutations introduced into RamC.