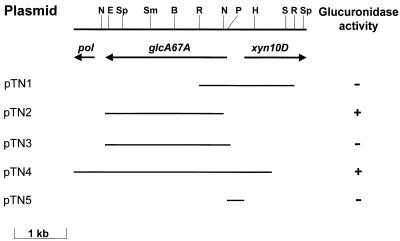
FIG. 1.
Physical map of agu67A locus. The positions of the cleavage sites for the following restriction enzymes are indicated: BamHI (B), EcoRI (R), EcoRV (E), HindIII (H), NcoII (N), PstI (P), SalI (S), SmaI (Sm), and SphI (Sp). The arrows show the extents and orientations of the genes encoding GlcA67A (agu67A), Xyn10D (xyn10D), and truncated DNA polymerase II (pol). Plasmid pTN4 was derived from recombinant phage, and pTN3, which encodes full-length GlcA67A, was a PCR product derived from pTN4. Plasmid pTN2, which encodes mature GlcA67A (residues 22 to 732), was derived from pTN3 by cloning the 2.1-kb NcoI-XhoI fragment into the expression vector pET32c. In pTN3 the α-glucuronidase gene was cloned into the pET expression vector pET28a, while pTN5 was derived by cloning a PCR product into the promoter probe vector pRG960SD. Glucuronidase activity refers to the presence of functional soluble enzyme activity in Escherichia coli strains carrying the plasmids.