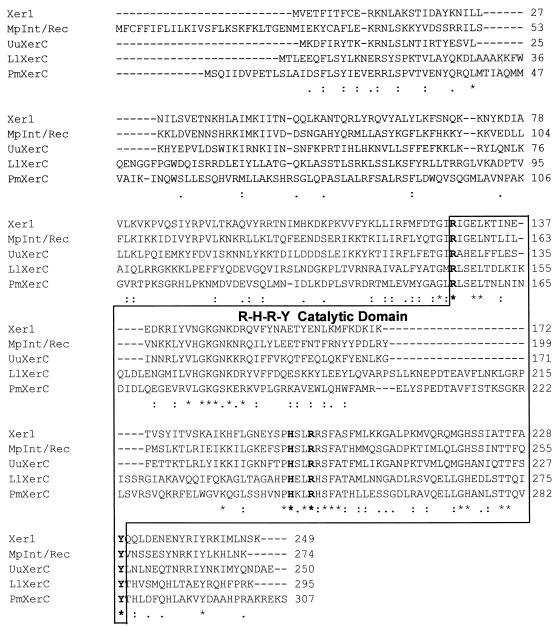
FIG. 6.
Amino acid alignment of Xer1 from M. agalactiae type strain PG2 with XerC recombinases from M. pulmonis (MpInt/Rec), U. urealyticum (UuXerC), Proteus mirabilis (PmXerC), and Lactobacillus leichmannii (LlXerC). The boxed C-terminal sequences form the catalytic domain that includes the four amino acids (R, H, R, and Y, shown in boldface type) that are invariant for the λ integrase family of recombinases. Dashes indicate gaps introduced to optimize alignment, and asterisks indicate positions which have a single fully conserved residue. Colons and dots indicate that one residue of the strong and weak groups (as defined by ClustalW, version 1.8) is fully conserved.