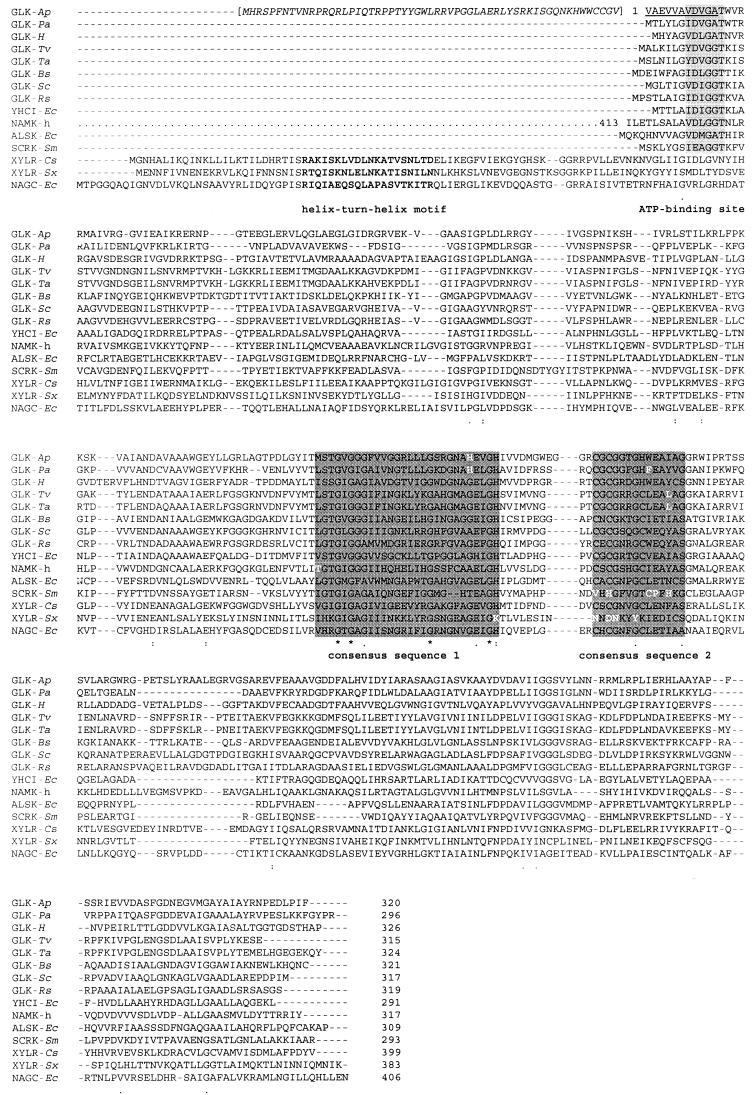
FIG.4.
Multiple sequence alignment generated by Clustal X (55) of deduced amino acid sequences of ATP-GLK from A. pernix and putative archaeal GLKs with selected members of the ROK family, which comprises sugar kinases and repressors (6, 56). Deduced amino acid sequences are shown for proteins from the following organisms (accession numbers given in parentheses): GLK-Ap, GLK from Aeropyrum pernix, with the N-terminal extension of ORF APE2091 given in italic letters and brackets and with the N terminus according to N-terminal sequencing underlined; GLK-Pa, putative GLK from Pyrobaculum aerophilum (PAE3437); GLK-H, putative GLK from Halobacterium sp. strain NRC1 (Q9HMA7); GLK-Tv, putative GLK from Thermoplasma volcanicum (Q97AS0); GLK-Ta, putative GLK from Thermoplasma acidophilum (Q9HJY6); GLK-Bs, GLK from Bacillus subtilis (P54495); GLK-Sc, GLK from Streptomyces coelicolor (P40184); GLK-Rs, GLK from Renibacterium salmoninarum (Q53165); YHCI-Ec, product of E. coli yhcI (P45425); NAMK-h, N-acetylmannosamine kinase domain of the bifunctional enzyme UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (residues 413 to 722) (Q9Y223); ALSK-Ec, allokinase from E. coli (P32718); SCRK-Sm, fructokinase from Streptococcus mutans (Q07211); XYLR-Cs, xylose repressor from Caldicellulosiruptor sp. (P40981); XYLR-Sx, xylose repressor from Staphylococcus xylosus (P27159); and NAGC-Ec, N-acetylglucosamine repressor from E. coli (P15301). The helix-turn-helix-motif found in the repressors (56) is marked in boldface; the ATP-binding (3, 14) site found in the kinases is shaded gray; and the two consensus sequences proposed, [LIVM]-x(2)-G-[LIVMFCT]-G-X-[GA]-X-G-X(3-5)-[GATP]-X(2)-G-[RKH] (19) and C-X-C-G-X(2)-G-X-[WILV]-E-X-[YFVIN]-X-[STAG] (10), are shaded dark gray.