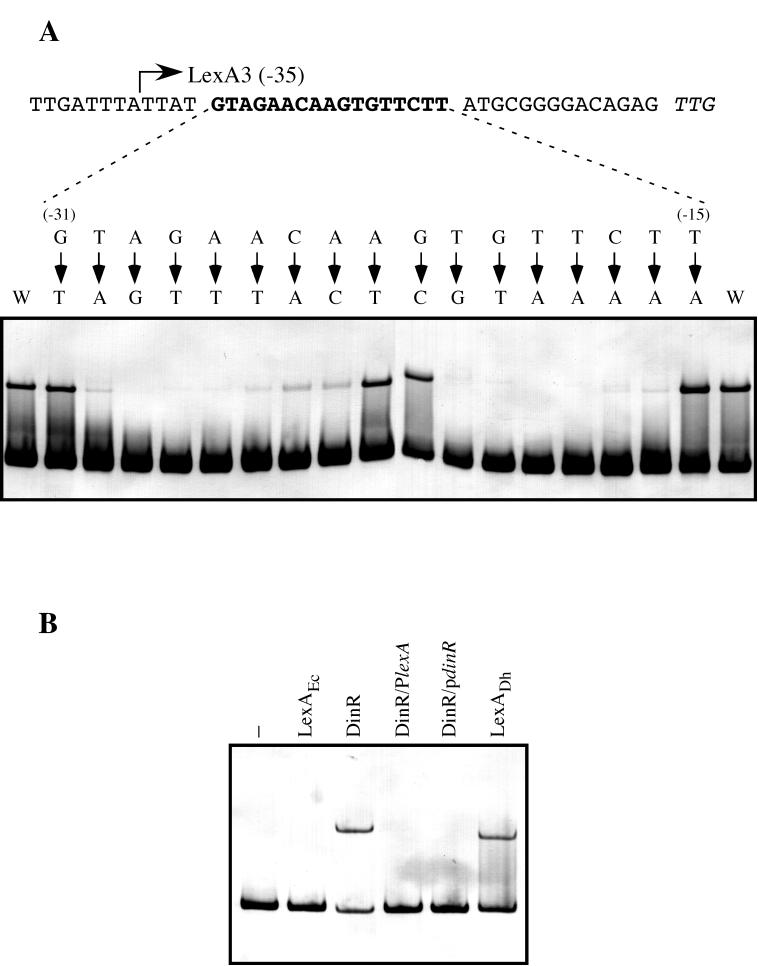
FIG. 4.
(A) Determination of the specific LexA binding site in the D. ethenogenes lexA promoter. Directed mutagenesis via PCR was used to change single nucleotides in the region from −31 to −15 (bold type) relative to the putative start codon of the D. ethenogenes lexA gene (italics). Electrophoretic mobility shift assays were used to determine the affinity of the purified LexA protein for the resulting digoxigenin-labeled DNA fragments. Arrows point to the nucleotide used to replace the native nucleotide. The relative positions of nucleotides with respect to the translation initiation codon are indicated in parentheses. The wild-type LexA3 fragment was used as a positive control (W). (B) Bacillus subtilis DinR recognizes the D. ethenogenes LexA binding site. Digoxigenin-labeled LexA1 fragment was incubated in the absence (−) or presence of 25 ng of either purified E. coli LexA (LexAEc), pure B. subtilis DinR (DinR), or D. ethenogenes LexA (LexADh). Furthermore, the LexA1 fragment and the DinR protein were incubated in the presence of a 100-fold molar excess of either the B. subtilis dinR (DinR/PdinR) or D. ethenogenes lexA (DinR/PlexA) promoter sequence.