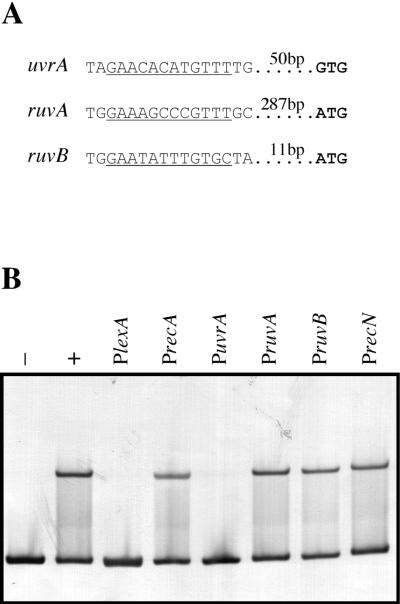
FIG. 5.
(A) Upstream sequences of D. ethenogenes uvrA, ruvA, and ruvB genes presenting putative LexA binding sites. Potential LexA binding sites are underlined, and the putative translation initiation codons are depicted in bold letters, with the distance between them indicated. (B). Electrophoretic mobility shift assay of the LexA1 fragment incubated with purified D. ethenogenes LexA in the presence of different DNA competitors. LexA1 fragment was incubated at 30°C for 30 min in the absence (−) or presence (+) of pure LexA protein. Simultaneous reactions contained the LexA1 fragment, D. ethenogenes LexA protein, and one of six unlabeled DNA competitors (100-fold molar excess) containing the promoter regions and potential LexA binding sites of genes lexA (PlexA,), recA (PrecA), uvrA (PuvrA), ruvA (PruvA), ruvB (PruvB), and recN (PrecN).