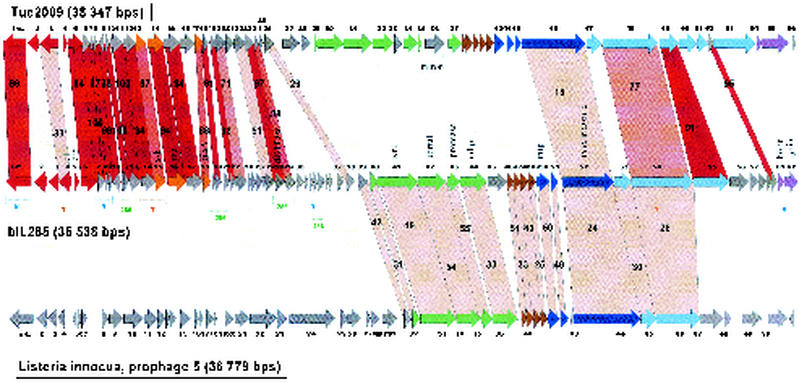
FIG. 6.
Alignment of the genetic maps from the temperate L. lactis phage Tuc2009 with L. lactis prophage bIL285 and L. innocua prophage 5. The open reading frames are color coded according to their predicted function as in Fig. 1. Selected genes from bIL285 are annotated. Genes encoding proteins that showed significant amino acid sequence similarity are linked by red shading, and the percentage of amino acid identity is indicated. The degree of amino acid identity (>90, >80, >70, or <70%) is reflected in the color intensity of the red shading. The brackets under the bIL285 gene map demarcate the patchwise sequence sharing of bIL285 genes with other lactococcal phages (B, BK5-T, T, TP901-1; 286, bIL286), possibly identifying genes whose proteins have to interact in a common function (see the text).