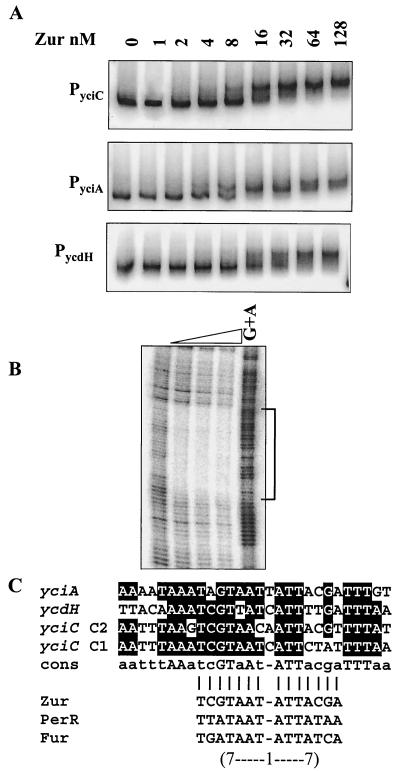
FIG. 5.
Interaction of Zur with its target operons. (A) EMSA of Zur binding to the promoter regions of yciC, yciA, and ycdH. Zur protein (concentrations indicated in nanomolar monomer) was incubated with 32P-end-labeled fragments and analyzed by 6% polyacrylamide gel electrophoresis. (B) DNase I footprinting analysis of Zur binding to the yciA promoter region. Zur protein was incubated with a 32P-end-labeled DNA fragment, and a G+A chemical cleavage ladder was included for localization of the binding sites. The concentration of Zur (monomer) in each reaction was (from left to right) 0, 20, 40, or 100 nM. (C) Alignment of the Zur-protected DNA regions in the promoters of yciA, yciC, and ycdH. The bottom part of panel C compares the core region protected by Zur (a 7-1-7 inverted repeat motif designated a Zur box) with similar operator sites recognized by the Fur homolog PerR and by Fur (1, 17).