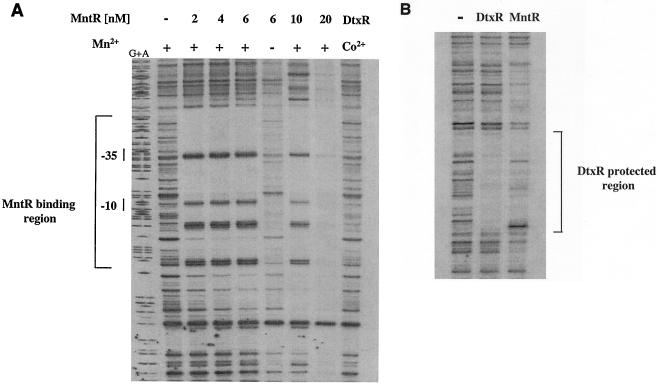
FIG. 5.
DNase I footprinting experiments. (A) Footprinting studies were done by using the DNA fragment used in the gel mobility shift assays. Mn2+ and Co2+ were used at 100 μM concentration, and native MntR was used at the concentrations indicated. The use of Co2+ to activate DtxR has been described previously (36). The bracket identifies the region protected by MntR from DNase I digestion, and −10 and −35 show the locations of the P1 promoter elements. G+A indicates the DNA sequencing ladder. (B) Footprinting experiments were done with a DNA fragment carrying the tox promoter region (43). MntR was used at 6 nM, and DtxR was present at 100 nM. The bracket indicates a 30-bp region protected by DtxR from DNase I digestion (43, 47). Co2+ and Mn2+ were used at 100 μM to activate DtxR and MntR, respectively.