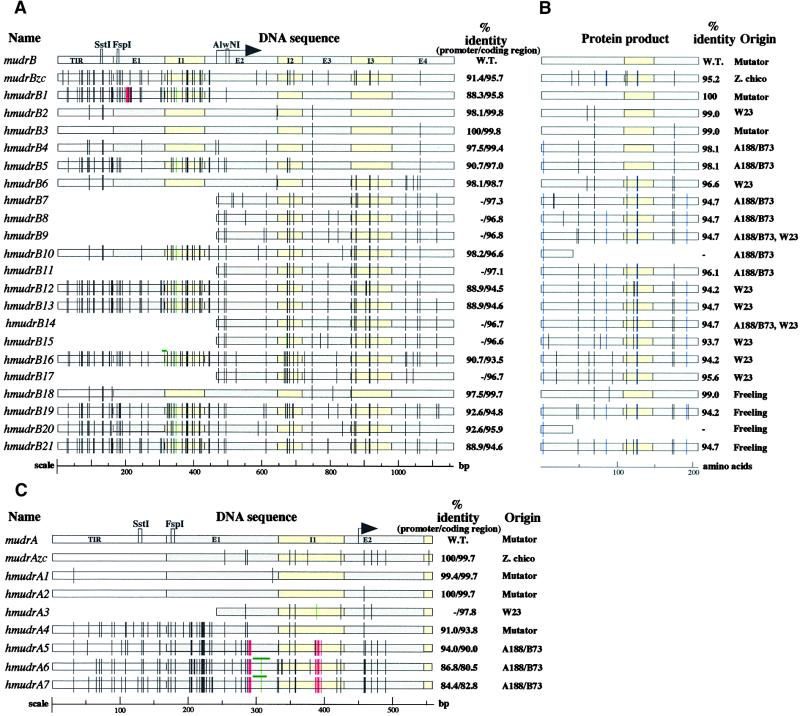
Figure 2.
Sequence Analysis of MuDR Homologs.
(A) Alignment and analysis of sequenced mudrB homologs. Wild-type (W.T.) mudrB is the top line, followed by the previously reported mudrB from the land race Zapalote chico, mudrBzc; the remaining sequences were recovered in this study, and their origins are listed in the column at right (B). Note that some homolog types were recovered from independent inbred lines. The TIR regions, introns, and exons are drawn to scale; the arrow indicates the translation initiation start site. Observed DNA mutations are indicated by vertical bars or boxes according to a color code: black, single base pair substitutions; green, insertions; red, deletions. As an internal control to account for potential point mutations that might have been derived as a result of clone propagation in Escherichia coli, six full-length wild-type mudrB genes were sequenced from an active Mutator line; these genes had the same sequence as the original MuDR isolate (Hershberger et al. 1995). In addition, single base DNA substitutions have been noted previously in three MuDR clones (Hershberger et al., 1995) and in the independently isolated MuA2 element (James et al., 1993).
(B) The MURB protein homologs encoded by each sequenced gene in (A) are shown as retaining intron 3 (207–amino acid form of MURB), with black bars corresponding to conservative substitutions and blue bars representing nonconservative substitutions. Truncated protein products are shown for hmudrB10 and hmudrB20, which share the same mutation creating a stop codon.
(C) Graphic alignment of sequenced regions corresponding to the mudrA gene. Annotations are the same as given in (A).