Figure 2.
The N. benthamiana PCNA Promoter Is Active in Cycling Cells.
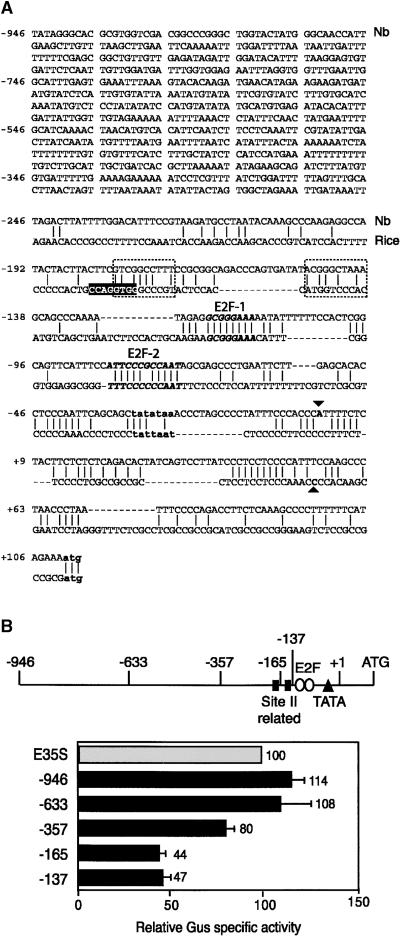
(A) The sequence of the longest N. benthamiana (Nb) PCNA promoter fragment is shown. The transcription start site (▾) was determined by S1 nuclease protection (data not shown). The proximal sequences of the N. benthamiana and rice (Suzuka et al., 1991) PCNA promoters are compared. The TATA boxes and ATG start codons of the two genes are shown in bold lowercase letters. The G-box (white letters), the site II (dotted boxes) elements, and the transcription start site (▴) of the rice promoter (Kosugi et al., 1995) are marked. The regions of the N. benthamiana promoter that show limited homology with site II sequences are included in the dotted boxes. Two sequences that display strong homology with the E2F binding consensus and are conserved between the two promoters are shown in bold italic type.
(B) A linear diagram of the N. benthamiana promoter indicating the site II–related elements, the E2F consensus elements, the TATA box, the transcription start site (+1), and the ATG. The end points of the 5′ deletions are marked. The relative reporter activities supported by the N. benthamiana promoter fragments in transiently transfected BY-2 cells are shown below. For each experiment, the activity of the enhanced Cauliflower mosaic virus 35S (E35S) promoter was set to 100 and the activities of the PCNA:uidA constructs were standardized against it. In these experiments, an average of 722 × 103 light units/μg protein was detected for the E35S:uidA construct, whereas an average of 915 light units/μg protein was detected for a promoterless uidA cassette. Gus, β-glucuronidase. The error bars represent two standard errors.