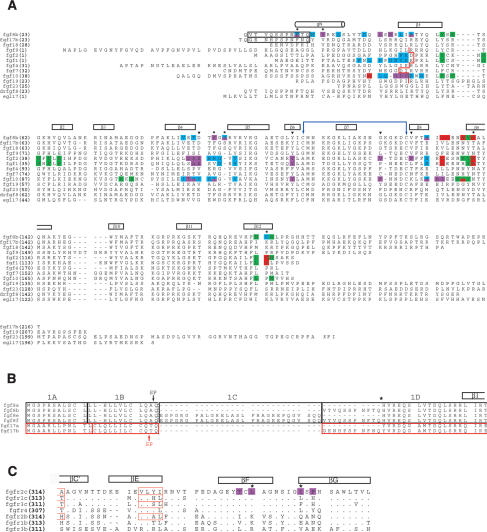
Figure 2.
Structure-based sequence analysis of the FGF8-FGFR mode of receptor-binding specificity. (A) Structure-based sequence alignment of selected FGFs. Predicted signal sequences have been omitted from the alignment. Residue numbers are in bold print to the left of the sequence alignment. FGF8 and FGF17 are numbered according to the “b” isoform of these ligands. The alternatively spliced N-terminal regions of FGF8b and FGF17b that are absent in FGF8a and FGF17a, respectively, are enclosed in a black box. The location of the secondary structural elements of FGF8b are shown on top of the sequence alignment. The N-terminal boundary of the β1 strands of other structurally characterized FGFs are marked by a red line. The N-terminal boundaries of FGF1, FGF2, and FGF10 have been assigned using the receptor-bound form of these ligands (PDB identification codes 1DJS, 1EVT, and 1NUN, respectively). The cysteine residues in FGF8b that form the disulfide bridge are marked with a violet double-headed arrow. A dash represents a gap introduced to optimize the alignment. FGF residues that interact with receptor D2, linker, the constant region of D3, and the alternatively spliced region of D3, are colored green, red, cyan, and purple, respectively. The six FGF8b residues that play a particularly important role in determining the unique mode of FGF8b receptor-binding specificity are marked with an asterisk. The FGF8b residues that correspond to the FGF1, FGF2, and FGF10 residues that form the βC′-βE loop-interacting hydrophobic patch of these ligands are indicated by black triangles. The sequence of Danio rerio FGF8b (drfgf8) and C. elegans EGL17 are also shown to highlight the evolutionary conservation of the unique mode FGF8b-FGFR-binding specificity. (B) Sequence alignment of the alternatively spliced N-terminal region of the human FGF8 and FGF17 isoforms. The amino acid sequences in the FGF8 and FGF17 proteins have been boxed according to the exons from which they are encoded: FGF8 exons are boxed in black and FGF17 exons are boxed in red. Note that the “a” isoforms of FGF8 and FGF17 are generated by the use of an internal splice-acceptor site in exon 1D (indicated by asterisk). The exon boundaries for FGF8 and FGF17 were obtained from Gemel et al. (1996) and Xu et al. (1999), respectively. The predicted signal peptide cleavage sites for FGF8 and FGF17 (Tanaka et al. 1992; Hoshikawa et al. 1998) are indicated by a black and red arrow, respectively. (C) Structure-based sequence alignment of the alternatively spliced, C-terminal half of D3 from human FGFRs. The location and length of the β strands are shown on top of the sequence alignment. A period indicates sequence identity to FGFR2c. A dash represents a gap introduced to optimize the alignment. FGFR2c residues that form the D3 hydrophobic groove and that interact with FGF8b are colored purple. Two key constituents of the groove are indicated with asterisks. The βC′ and βE strands of FGFR1c, FGFR2c, FGFR3c, and FGFR2b from previous FGF-FGFR structures (PDB identification codes: 1EVT, 1DJS, 1RY7, and 1NUN, respectively) are indicated by red boxes.