FIG. 1.
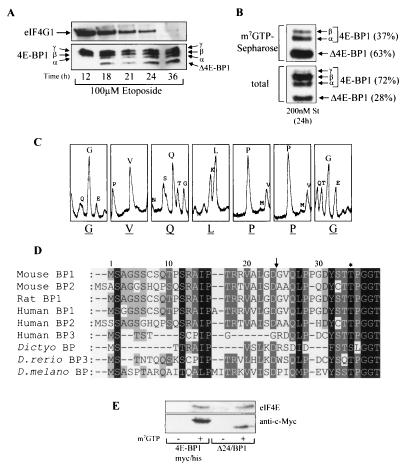
Identification of the caspase-dependent cleavage of 4E-BP1. (A) Swiss 3T3 cells were treated with etoposide for the times indicated. The samples were analyzed for total eIF4G1 (top) and 4E-BP1 (bottom). The α-, β-, and γ-species of 4E-BP1 and cleavage product Δ4E-BP1 are labeled. (B) Swiss 3T3 cells were treated with staurosporine (St) for 24 h, and extracts were subjected to affinity chromatography on m7GTP-Sepharose (see Materials and Methods). The percentages of uncleaved 4E-BP1 (γ, β, and α) and cleaved (Δ4E-BP1) 4E-BP1 bound to eIF4E (top) and total levels present within cell extracts (bottom) were quantified by densitometry (National Institutes of Health Image, version 1.61). (C) Δ4E-BP1 (isolated as described for panel B) was excised from the membrane and subjected to Edman degradation, yielding N-terminal sequence GVQLPPG. (D) N-terminal alignment of 4E-BPs. Arrow, cleavage site; ∗, phosphorylation site at Thr36 in mouse and rat 4E-BP1 and at Thr37 in human 4E-BP1. 4E-BP1 homologues from D. discoideum (Dictyo), D. rerio, and D. melanogaster (D. melano) are also shown. (E) Cell lysates prepared from serum-starved HEK293 cells overexpressing either 4E-BP1myc/his or Δ24/BP1 (see Fig. 2A for details of these mutants) were subjected to affinity chromatography using either m7GTP-Sepharose (+) or Sepharose (CL-4B; −). The levels of purified recombinant 4E-BP1 and eIF4E were analyzed.