Abstract
The “one-gene, one-protein” rule, coined by Beadle and Tatum, has been fundamental to molecular biology. The rule implies that the genetic complexity of an organism depends essentially on its gene number. The discovery, however, that alternative gene splicing and transcription are widespread phenomena dramatically altered our understanding of the genetic complexity of higher eukaryotic organisms; in these, a limited number of genes may potentially encode a much larger number of proteins. Here we investigate yet another phenomenon that may contribute to generate additional protein diversity. Indeed, by relying on both computational and experimental analysis, we estimate that at least 4%–5% of the tandem gene pairs in the human genome can be eventually transcribed into a single RNA sequence encoding a putative chimeric protein. While the functional significance of most of these chimeric transcripts remains to be determined, we provide strong evidence that this phenomenon does not correspond to mere technical artifacts and that it is a common mechanism with the potential of generating hundreds of additional proteins in the human genome.
It is now well established that the genetic complexity of an organism cannot be directly extrapolated from its gene number. Alternative splicing, for example, dramatically increases protein complexity. Extensive EST sequencing (for review, see Modrek and Lee 2002) and exon-junction microarrays (Johnson et al. 2003) have demonstrated that alternative splicing occurs in most human genes; thus the 20,000 to 25,000 genes currently estimated in the human genome (The International Human Genome Consortium 2004) may potentially encode a much larger number of different proteins. In addition to alternative splicing, other widespread mechanisms exist that contribute to genome complexity. These include RNA editing (Athanasiadis et al. 2004; Blow et al. 2004), trans-splicing (Takahara et al. 2005), alternative transcription start sites (Hashimoto et al. 2004), and alternative polyadenylation transcription termination sites (Beaudoing and Gautheret 2001).
Here we investigate another phenomenon that may also contribute to generate additional complexity in the genome. Several reports published within the last decade (for review, see Akiva et al. 2006 in this issue) describe a similar phenomenon: Two adjacent genes in the same orientation that are usually transcribed independently are occasionally transcribed into a single RNA sequence whose splicing product encodes a protein including coding exons from the two genes (see Supplemental Fig. 1). We refer here to this RNA product as a Transcription Induced Chimera or TIC, since we hypothesize that run-off transcription is the most likely mechanism involved in its generation (see Discussion). TICs are different from polycistronic operons in prokaryotes, in which a single transcript translates into different proteins—but no chimeric proteins or independent transcripts are produced. Here we attempt to estimate the frequency of Transcription Induced Chimerism in the human genome and thus assess to what extent it could constitute an additional mechanism to generate protein diversity. Our estimation is based both on a genome-wide survey of existing EST sequences and on the systematic verification of de novo computational predictions of chimeric transcripts obtained in the regions selected within the ENCODE project (The ENCODE Project Consortium 2004). Results in the ENCODE regions suggest that at least 4% to 5% of the tandem genes in the human genome can be occasionally transcribed into a single RNA sequence with the potential of encoding a chimeric protein sequence.
Results
Genome-wide survey of chimeric transcripts supported by EST sequences
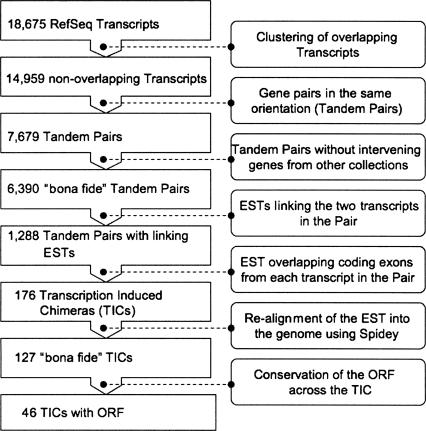
To obtain an initial estimate of the frequency of Transcription Induced Chimeras (TICs), we first compared Expressed Sequence Tags (ESTs) with the set of known human genes in the RefSeq database (see Fig. 1). The coordinates of 18,675 RefSeq transcripts were obtained from the University of California at Santa Cruz (UCSC) Browser. When several RefSeq transcripts overlapped, only the longest was considered. This resulted in 14,959 non-overlapping genes, of which 7679 were identified as tandem pairs (pairs of adjacent genes encoded in the same orientation). We further considered only those pairs for which there was no evidence of an intervening gene from other gene collections (those in the UCSC browser tracks from the Vertebrate Genome Annotation database (VEGA; http://vega.sanger.ac.uk/) and “known genes”), bringing the number down to 6369 tandem pairs. The coordinates of the ESTs on the human genome sequence were also obtained from the UCSC Browser. Comparing ESTs with RefSeq coordinates we found that in 1288 of the above tandem pairs, at least one EST reached across the boundaries of the two genes in the pair. In 176 of these, the chimeric EST covered at least one coding exon from each of the tandem genes. Using the program Spidey, which takes into account consensus splice-site boundaries, these cases were further confirmed through realignment on the human genome sequence, and 127 cases remained that had an alignment >100 bp with at least 95% identity and one intron spliced out across the tandem genes (see Fig. 1 for a detailed flowchart of the entire protocol). This set included four of the 13 previously described TICs (see Akiva et al. 2006; Table 1); those involving the genes GALT (RefSeq id NM_000155) and IL11RA (NM_004512) (Magrangeas et al. 1998), CYP2C18 (NM_000772) and CYP2C19 (NM_000769) (Zaphiropoulos 1999), VPS72 (NM_005997) and TMOD4 (NM_013353) (Cox et al. 2001), and PPAN (NM_020230) and P2RY11 (NM_002566) (Communi et al. 2001). For the remaining nine published chimeras, there was either no RefSeq entry for at least one of the tandem genes, or there was no EST evidence of chimerism.
Figure 1.
Flowchart depicting the protocol used to infer EST-supported TICs.
Table 1.
Functional associations between components of RefSeq transcriptional chimeras supported by ESTs
|
No. RT-PCR positives
|
Physical associationa
|
Functional associationb
|
Related gene fusionsc
|
|||||
|---|---|---|---|---|---|---|---|---|
| Chimera | No. ESTs | Same/orth. | COG | Same/orth. | COG | Fusion | Organism | |
| NME1–NME2 | 4 | 7 | Yes | Yes | TXN-NDK(3) | HS, MM | ||
| NDK-DSCAML1 | MM | |||||||
| ELA3A–ELA3B | 1 | — | Yes | Yes | SMO/FZD-Trypsin like | HS | ||
| Fibrillin-trypsin like | HS | |||||||
| Scavenger receptor-trypsin like | HS | |||||||
| BHMT2–BHMT | 1 | — | Yes | Yes | BHMT-Methionine synthase | HS, MM, CE | ||
| ACAD10–ALDH2 | 1 | 1 | No | Yes | No | Yes | ALDH-ADH | Prokaryotes |
| PRODH-ALDH | Prokaryotes | |||||||
| HAD-APH-ACAD | HS, MM, CE | |||||||
| ASL–RCP9 | 1 | — | No | Yes | No | Yes | NAD oxidoreductase-RPC17 | CE |
| GIMAP2–GIMAP1 | 1 | No | No | Yes | ||||
| PLCXD2–PHLDB2 | 2 | 1 | No | No | Yes | |||
| DTX2–PMS2L5 | 2 | 1 | No | Yes | No | Yes | 5′–3′ exonuclease-helicase | Prokaryotes |
| SENP3–EIF4A1 | 1 | — | No | Yes | No | Yes | ||
| HIF1A–SNAPC1 | 1 | — | No | No | No | Yes | ||
| MIA–RAB4B | 1 | — | No | No | No | Yes | ||
| SLC2A11–MIF | 1 | — | No | No | No | Yes | MIF-PGLYRP | MM |
| HSPH1–PREI3 | 2 | 1 | No | No | No | Yes | ||
| SNTB2–VPS4A | 1 | — | No | No | No | Yes | ||
| ATP6V0C–CGI-14 | 1 | — | No | No | No | Yes | ||
| TMPIT–STYXL1 | 2 | 2 | No | No | No | Yes | ||
| FPGT–TNNI3K | 3 | 4 | No | No | No | Yes | ||
Physical interactions between components or their orthologs (Same/orth.) or proteins within Clusters of Orthologous Sequences (COG) were retrieved from BIND databases (http://bind.ca/). These associations can be either direct, or as part of multi-subunit complexes. For interactions for which only yeast two-hybrid data are available, only those with high-confidence were considered
Functional associations between chimera components or proteins within COGs were inferred from the STRING server (http://string.embl.de/) and from the KEGG metabolic pathways database
Previously described fusions involving one of the two partners of the tandem chimeras are shown on the first column. Information for gene fusions was retrieved from STRING (http://string.embl.de/). (HS) Homo sapiens; (MM) Mus musculus; (CE) Caenorhabditis elegans
In several cases, the EST-supported TICs included additional exons encoded in the “intergenic” sequence between the two fused genes (26 out of 127, 21%), but the most common arrangement was the splicing from the penultimate exon in the upstream gene to the second exon in the downstream gene (32 out of 127, 27%)—perhaps as a means to escape the stop codon in the terminal exon of the upstream gene. See Akiva et al. (2006) for a more detailed discussion of the anatomy of TICs. In 45 additional cases, the chimera included the terminal exon of the upstream gene. Interestingly, in 23 of those cases, splicing internal to the coding fraction of the exon avoided the stop codon. On average, 419 bp from the upstream gene and 537 bp from the downstream gene are covered by the ESTs that support the TIC.
In 46 cases of the 127, the coding frame of the downstream gene was maintained across the chimeric junction. This is only marginally higher than the one-third expected by chance. The complete set of detected TICs is available at http://genome.imim.es/datasets/chimeras2005, and Table 1 lists some of the most interesting cases (see also Discussion).
The majority of the TICs supported by ESTs are supported by a single EST (90 out of 127). However, the number of supporting ESTs is not larger for the TICs corresponding to the four known as compared to the number of ESTs supporting the novel TICs. The four known chimeras are supported by just one EST. Therefore, low EST copy number may not reflect underlying artifacts but, rather, restricted expression patterns.
In this regard, we have experimentally investigated the physiological presence of a subset of the novel EST-supported TICs by RT-PCR on 12 different tissues (see Methods). Of the 46 cases in which the ORF was conserved, we tested the 32 cases in which the TIC did not include additional “intergenic” exons. Of these, 11 (34%) yielded specific amplification products in at least one tissue. The lower success rate in our case than in the experiments of Akiva et al. (2006) may be caused by the fact that we used oligo(dT) priming to generate the cDNA libraries (see Methods), while Akiva et al. (2006) used random priming in addition. Random primers are likely to be more effective in recovering longer mRNA sequences such as those produced by the chimeric transcripts. As expected, the positive verification rate was higher for those TICs supported by at least two ESTs (three out of seven, 43%), or three ESTs (three out of three, 100%). Most—but not all (12 out of 21)—of the EST-supported chimeras that failed RT-PCR verification were supported by ESTs from libraries not corresponding to the 12 tissues used in the RT-PCR experiments. Among the positive cases, the average number of tissues in which the TICs were expressed was 2.5, far below the seven to eight positive tissues out of 12 tested for known mammalian genes (Reymond et al. 2002b; Waterston et al. 2002), but comparable to that obtained for novel human genes identified using the recently released chicken genome as reference (Castelo et al. 2005).
A similar protocol was followed with the mouse genome, leading to similar results (see Supplemental material).
Identification of novel chimeric transcripts by experimental verification of computational predictions
The above genome-wide analysis suffers from the limitation that the RefSeq collection of genes—based on mRNA evidence—includes only a fraction of all human genes. This leads to the misidentification of tandem genes, and to an underestimation of the frequency with which EST sequences are suggestive of tandem chimeras. Also contributing to the underestimation is the fact that we have used only one representative transcript from each set of overlapping transcripts. To address this limitation, we have performed a more detailed analysis in the 1% of the human genome targeted within the ENCODE project (The ENCODE Project Consortium 2004). The 44 regions selected within this project are being exhaustively scrutinized, and are rich in genes. For this analysis, in addition to transcripts in RefSeq, we also considered genes in the VEGA database and the “known genes” track on the UCSC Browser, the latter based on SWISS-PROT, TrEMBL, known mRNAs, and also RefSeq. A total of 992 unique coding sequences were identified from these gene sets in the ENCODE regions. Transcripts without complete ORFs, or with ORFs lacking a starting methinonine or a terminating stop codon were removed, leaving 594 transcripts. The comparison of this number with the 14,959 coding RefSeq transcripts on which our genome-wide analysis was based emphasizes the deeper transcript coverage available to us for the analysis of the ENCODE regions. These transcripts were clustered into 321 non-overlapping gene loci, in which 165 tandem gene pairs were identified. These were compared against ESTs, and after detailed inspection, six tandem pairs were found (3.6%), in which EST sequences maintaining the ORF linked coding exons from transcripts belonging to the two genes within the tandem. Only one of these chimeric transcripts had been previously discovered through our genome-wide analysis of RefSeq genes.
Because of the deeper transcript coverage of the ENCODE regions, we believe that this is a more realistic estimate of tandem chimeric transcription than that obtained in the genome-wide survey, based on RefSeq genes. However, since EST libraries capture only a fraction of the transcription and splicing diversity in the human genome (Kapranov et al. 2002; Johnson et al. 2003), it is possible that the frequency of TICs provided here is still an underestimate. Therefore, to obtain an estimate that is less biased toward currently available transcript data, we have systematically tested tandem genes for potential chimerism by means of RT-PCR. We have proceeded in the following way: We extracted the genome sequence from the 5′-end of the upstream gene to the 3′-end of the downstream gene for each tandem pair, and then forced the program geneid (Parra et al. 2000) to predict a single complete gene along this sequence (see Methods). Predictions overlapping the coding exons of the two underlying tandem genes were obtained in 126 cases (out of 165), of which 92 were randomly selected for RT-PCR verification on RNAs from 24 human tissues (see Methods). In each of these cases, primer sequences were chosen from pairs of predicted internal exons, each predicted exon overlapping a real exon from each of the underlying tandem genes. Three of the 92 cases yielded positive results after RT-PCR and sequencing of the amplimer. Figure 2 shows these cases. Two of them corresponded to tandem chimeras already supported by ESTs—a number consistent with the rate of RT-PCR verification of EST-supported chimeras that we had previously observed—but the third one was novel. Altogether, we have evidence for seven chimeric transcripts out of 165 tandem pairs. The actual number of TICs could still be higher, because we have only tested about three-fourths of the TICs predicted computationally. In summary, our analysis indicates that between 4% and 5% of the tandem genes in the ENCODE regions could be encoding a chimeric protein.
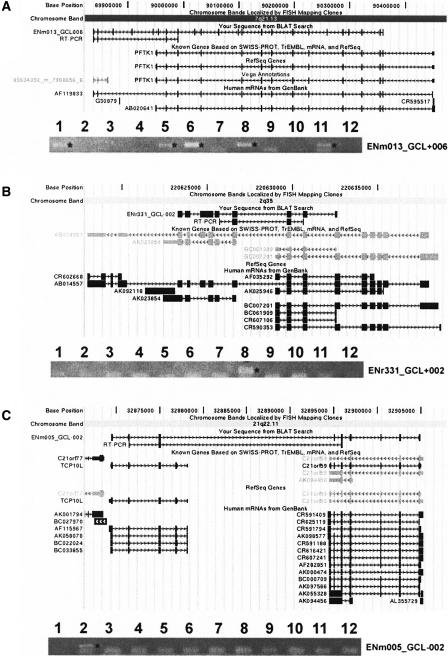
Figure 2.
The three computationally predicted chimeras in the ENCODE regions verified by RT-PCR. (A) chimera ENm013_GCL+006; (B) chimera ENr331_GCL+002; (C) chimera ENm005_GCL-002. Only one had known chimeric mRNAs before our experiments. One of the chimeras—ENm005_GCL-002—still lacks known mRNAs. The results of the RT-PCR validation in 12 of the 24 tissues tested are shown. Asterisks mark positive amplimers. The tissues are (1) brain, (2) heart, (3) kidney, (4) spleen, (5) liver, (6) colon, (7) small intestine, (8) muscle, (9) lung, (10) stomach, (11) testis, and (12) placenta. Chimeras were tested in 12 additional tissues (not shown here).
Discussion
Our analysis suggests, therefore, that Transcription Induced Chimerism could contribute to generate additional protein complexity. Indeed, extrapolating the results from our analysis of the ENCODE regions to the entire human genome would imply the existence of hundreds of additional protein sequences generated by chimerism of tandem genes. Certainly, it is still unclear to what extent the ENCODE regions are representative of the entire genome, and the numbers that we have obtained with this set may be too small to make the extrapolation statistically significant. On the other hand, however, our analysis of the ENCODE regions suffers from several limitations that are likely to result in an underestimation of Transcription Induced Chimerism. First, it is becoming increasingly evident that the EST sequences capture only a fraction of the transcript diversity of the human genome (Kapranov et al. 2002; Cheng et al. 2005). Indeed, most TICs are supported by a single EST, indicating that they are rare events, and therefore likely to be under-represented in the current EST data sets. Assuming that the number of ESTs per TIC follows a Poisson distribution, we estimate that for at least 25% of the real TICs, EST support will not exist (see Supplemental material). Second, our approach to predict and validate chimeric transcripts faces the following limitations: (1) we relied on ab initio computational predictions, which are still inaccurate; (2) we verified by RT-PCR only a pair of exons from each predicted chimera; and (3) we used cDNAs from a limited set of tissues to test the transcription potential. Therefore, it cannot be ruled out that the fraction of tandem genes that produce TICs is much higher than 4% to 5%—a number that should be taken only as a lower bound estimate.
Regarding the mechanism through which TICs are generated, we favor the hypothesis that they result from run-off transcription of the upstream gene, which then stops at the Transcription Termination Site of the downstream gene. This results in a chimeric transcript, which is subsequently spliced into a chimeric mRNA. This is, otherwise, the mechanism commonly postulated to explain the previously documented cases (see, e.g., Thomson et al. 2000; Pradet-Balade et al. 2002; Poulin et al. 2003; and Roginski et al. 2004). An alternative possibility, however, is that TICs result from trans-splicing between the separate pre-mRNAs of the tandem genes. Few instances, however, have been reported of trans-splicing in mammals, and they often involve pre-mRNAs of the same gene (homotypic trans-splicing) (see Caudevilla et al. 1998; Takahara et al. 2005). Moreover, TICs often include intervening exons encoded in the intergenic space defined by the tandem genes (21% of the cases in our study of the ESTs supporting chimeric RefSeq genes). In these cases at least, trans-splicing appears as an unlikely mechanism. It can be argued that the tandem transcripts that we assume to correspond to two separate loci are actually alternative non-overlapping splice forms of a unique unrecognized larger locus, with alternative promoter and Transcription Termination Sites. In this interpretation, TICs would simply be additional splice forms of the loci. The issue here is whether the upstream gene has a legitimate transcription termination site, and the downstream gene has a legitimate promoter region. This would imply the existence of two (non-overlapping) independently regulated transcripts—encoded, therefore, by two separate loci. While again we lack the detailed molecular studies to conclude irrefutably one way or the other, we contend that most of the available evidence suggests that the majority of TICs are created from two bona fide genes in tandem. First, RefSeq transcripts usually have a corresponding full-length cDNA clone. Such clones are obtained by reverse transcription of the messenger RNA by oligo(dT) priming—which targets poly(A) tails often associated with the transcription termination site. Also, in support of this, we have computed the number of 3′-ESTs that terminate at the terminal exon of the downstream gene of the chimera-encoding tandem pair. Of the 127 TICs detected from the EST analysis, 83 of them (65%) have at least one 3′-EST terminating at the last exon of the upstream gene (with an average of 45 3′-ESTs per gene terminating at the last exon). In comparison, only in 10 cases (8%), we have at least one 3′-EST terminating at the first exon of the downstream gene (with an average of 1.25 ESTs per gene). Such largely different numbers would not be expected if the two tandem genes were instead non-overlapping alternative splice forms of a unique larger loci. On the other hand, Trinklein et al. (2003, 2004) (see http://genome.ucsc.edu/ encode for details), have investigated the activity levels of 643 promoter fragments predicted in the ENCODE regions based on high-throughput transient transfection luciferase reporter assays in a panel of 16 cell lines. In Table 2, we report the values of the activity levels for the seven detected TICs in the ENCODE regions. (To control for variation in transcription efficiency, a plasmid expressing Renilla luciferase is used; activity levels are then reported as normalized and log2-transformed averages on the firefly luciferase/Renilla luciferase ratio. See the aforementioned references for details). These values are assumed to be indicative of promoter activity when they are greater than three standard deviation units above the mean of the negative controls for each cell type (in parentheses in Table 2). According to this criterion, five upstream and four downstream genes in the seven tandem chimeras detected in ENCODE show positive promoter activity. Such similar numbers would again not be expected if the downstream gene were a mere alternative splice form of a single larger loci.
Table 2.
Chimeric transcripts found in the ENCODE regions
| ENCODE region | Chromosome | Gene 1 (upstream 5′) | Transcripts 1 | Promoter activity | Gene 2 (downstream 3′) | Transcripts 2 | Promoter activity | Supporting EST | RT-PCR |
|---|---|---|---|---|---|---|---|---|---|
| ENm009 | chr11 | TRIM6 | NM_001003818 | 45.88* | TRIM34 | NM_021616 | 242.41* | AB039903 | |
| NM_058166 | (1.11 ± 1.57) | NM_130389 | (1.06 ± 1.36) | ||||||
| HTC116 | MG63 | ||||||||
| ENr233 | chr15 | SERF2 | NM_005770 | 929.73* | HYpk | NM_016400 | 1.88 | AK000438 | |
| (1.05 ± 1.66) | (1.04 ± 1.45) | ||||||||
| Be2C | Panc1 | ||||||||
| ENm005 | chr21 | CRYZL1 | BC033023 | 82.11* | DONSON | NM_017613 | 9.90* | AL157441 | |
| (1.19 ± 2.23) | NM_145794 | (1.19 ± 2.23) | |||||||
| JEG3 | NM_145795 | JEG3 | |||||||
| ENr223 | chr6 | C6orf148 | NM_030568 | 230.36* | AC019205.8-001 | AK090984 | 115.18* | BM544101 | |
| bA257K9.4-002 | (1.05 ± 1.66) | (1.02 ± 1.12) | |||||||
| bA257K9.4-001 | Be2C | HMCB | |||||||
| ENm013 | chr7 | AC003076.1-001 | AC003076.1-001 | N/A | PFTK1 | NM_012395 | 2.43 | AF119833 | Brain |
| (1.22 ± 2.43) | Liver | ||||||||
| HepG2 | Colon | ||||||||
| Muscle | |||||||||
| Testis | |||||||||
| ENr331 | chr2 | Q96IW3 | BC007201 | 2.95 | Q9H8B3 | AK023854 | 11.61* | BI559709 | Muscle |
| (1.09 ± 1.41) | (1.19 ± 2.23) | BG912151 | |||||||
| G402 | JEG3 | ||||||||
| ENm005 | chr21 | C21orf59 | NM_021254 | 463.68* | TCP10L | NM_144659 | 1.27 | Heart | |
| NM_017835 | (1.01 ± 0.74) | (1.01 ± 0.74) | |||||||
| AK094456 | U87 | U87 | |||||||
| AK055328 |
Tandem chimeric transcripts in the ENCODE regions supported either by ESTs or RT-PCR of computational predictions, or both. Gene and Transcript identify the genes and transcripts involved in the TICs. Supporting ESTs are the ESTs supporting the TICs after our filtering protocol (see Methods). RT-PCR lists the tissues in which expression of the chimeric transcript has been detected in the RT-PCR experiment (see Fig. 2). Experimental validation of putative promoters by reporter assay is shown on the promoter activity column. The transcription start sites were predicted by assigning the 5′ end of each gene model as the transcription start site supported by full-length cDNAs. Relative reporter activity was determined by comparing the firefly luciferase/Renilla luciferase ratios of reporter constructs in 16 different cell lines (see Trinklein et al. 2003, 2004). The highest firefly luciferase/Renilla luciferase ratios values are shown together with the mean and the standard deviation of the negative controls for the corresponding cell line. Promoter fragments with a significant reporter activity (exceeding three times the standard deviation of all control fragments) are marked with an asterisk. The firefly luciferase/Renilla luciferase ratios from the promoter regions and the negative controls were obtained from the UCSC Browser.
It can also be argued that chimeric transcripts could be artifacts. However, the fact that EST-supported TICs are often independently verified by RT-PCR and that the TICs are spliced at canonical splice sites supports the argument against experimental artifacts. While we have not performed negative controls specific to this study, results from a previous study, where we attempted to prove expression of Conserved Noncoding Sequences (CNS) in human chromosome 21 (Dermitzakis et al. 2002), indicate that our RT-PCR protocol is very specific: We obtained no positive amplifications out of a set of 89 tested pairs of CNS.
On these grounds, we believe, therefore, that TICs are not technical artifacts. The issue remains, however, of their functional relevance. The possibility exists of widespread stochastic transcription and splicing, which would not necessarily translate into functional protein synthesis. In this regard, specific antibodies against the chimeric protein could be designed to demonstrate translation. Such protein products, however, could also result from stochastic translation void of functional significance. The functional relevance of tandem chimeric transcripts could be established beyond doubt only through the observation of phenotypic alterations after tandem disruption.
Existing data do not strongly support general functionality for the TICs identified here. For instance, the frequency with which the ORF is conserved across the TIC is only marginally higher than expected by chance. On the other hand, we have investigated the fraction of human TICs for which we have EST evidence of conservation in mouse. Indeed, conservation of tandem chimerism between human and mouse would be strongly suggestive of functionality. We have mapped the human TICs into the mouse syntenic regions and searched for mouse ESTs supporting the homologous mouse TIC. We have found conserved TICs in mouse for five of the 46 EST-supported human TICs with conserved ORFs. However, given the low number of ESTs that usually support TICs, it cannot be ruled out that, despite the absence of supporting ESTs, other human TICs are also present in mouse.
In any case, we believe that at least for a few TICs, specific functionality is a plausible hypothesis. Indeed, several of the tandem chimeric transcripts found in our survey correspond to proteins that, when expressed separately, are known to interact physically with each other or to belong to common biochemical pathways (see Table 1). At least for this set of chimeric transcripts, an analogy can be made to known gene fusions that link functionally related genes (Snel et al. 2000), in the sense that the chimeras with known or probable functional associations between their components are more likely to be functional (Enright et al. 1999; Marcotte et al. 1999). The NME1–NME2 chimera, for instance, is predicted to produce a protein with two catalytic sites for phosphate transfer. The nucleotide diphosphate kinases (NDK) NME1 and NME2 form homodimers that complex into tetramers or hexamers (Janin et al. 2000). Two NME modules within a single polypeptide might have functional implications in either of their two known activities, as enzymes that transfer phosphate to nucleosides, or as DNA-binding transcriptional regulators. NDK domains have been found fused to other domains (Sadek et al. 2001). Another example is that of the chimera formed by the genes for histone 2A and histone 3B, which are part of the nucleosome core complex (Luger et al. 1997). A composite protein with HIST1H2AI at its amino half and HIST1H3H at its carboxyl half may not disrupt the overall configuration relative to that of the histone octamer (Luger et al. 1997), and from that perspective it would be a functionally plausible chimera. Histone 2A-like domains are involved in at least one more gene fusion with a non-histone C-terminal component (Pehrson and Fried 1992). Additional chimeric transcripts involving fusions between functionally related genes include those of several duplicated genes coding for paralogs (ELA3A–ELA3B, BHMT–BHMT2, GIMAP2–GIMAP1) and also nonduplicated genes (ACAD10–ALDH2, PLCXD2–PHLDB2, DTX2–PMS2L5, HIF1A–SNAPC1). These chimeras are listed in Table 1, and their potential functional significance is discussed in more detail in the Supplemental material.
Whether functionally relevant or the product of stochastic transcription and splicing, widespread Transcription Induced Chimerism—as well as other phenomena that are becoming better understood as we scrutinize the genome sequence with unprecedented detail—are revealing a genome of unexpected complexity.
Methods
Data sets
Most of the data sets used in our analysis were obtained from the UCSC Genome Browser database (http://genome.cse.ucsc.edu/). The human NCBI33 (hg15, April 2003) assembly was used. The NCBI Reference Sequence (RefSeq; http://www.ncbi.nlm.nih.gov/RefSeq/) transcript coordinates were taken from the UCSC database refGenes.txt file. Coordinates of the EST mapping were obtained from the chrN_est, chrN_intronEst and chrN_mrna files. The ENCODE regions, and the associated gene annotation were also extracted from the UCSC Genome Browser database (http://genome.ucsc.edu/ENCODE/regions.html). They corresponded to the NCBI35 assembly.
Computational predictions of chimeric transcripts
The program geneid (Parra et al. 2000) was used to find ORFs in tandem chimeric ESTs, and to predict tandem chimeric proteins. For the latter, the genome sequence extending two tandem genes was given as input to geneid. If necessary, the sequence was reverse-complemented to guarantee that the tandem genes were encoded on the forward strand. While, by default, geneid predicts multiple genes in both strands, when used with the options -F and -W it predicts a single complete gene in the forward strand. We used geneid with these two options. In addition, geneid can use external information to guide the gene predictions (Blanco et al. 2003). Here we have chosen to provide to geneid the coordinates of the real coding exons of the underlying tandem genes as regions of similarity to existing proteins. This increases the likelihood of the underlying coding exons to be included in the final prediction, but it does not force their inclusion or prevent the inclusion of novel exons. Because the 3′-most exon of the upstream gene and the 5′-most exon of the downstream gene may be skipped in the chimeric transcript, they were not given to geneid.
Experimental transcript validation
Human cDNAs from 12 different tissues (brain, heart, kidney, spleen, liver, colon, small intestine, muscle, lung, stomach, testis, and placenta) were used to validate the chimeric transcripts supported by ESTs. Validation of the computational predicted chimeric transcripts was done using cDNAs from 24 tissues (the previous 12 tissues plus skin, peripheral blood cells, bone marrow, thymus, pancreas, mammary gland, prostate, fetal brain, fetal liver, fetal kidney, fetal heart, and fetal lung). The cDNAs were synthesized using 12 poly(A)+ RNAs from Origene, eight from Clemente Associates/Quantum Magnetics, and four from BD Biosciences as described (Reymond et al. 2002a,b). The relative amount of each cDNA was normalized by quantitative PCR using SybrGreen as intercalator and an ABI Prism 7700 Sequence Detection System. Putative tandem chimeric transcripts were assayed experimentally by RT-PCR as previously described (Reymond et al. 2002b; Guigó et al. 2003). To experimentally verify computationally obtained tandem chimeric transcripts, the primer sequences were designed in pairs of predicted internal exons, each predicted exon overlapping a real exon from each of the underlying tandem genes. Whenever possible, the selected exons overlapped the exon preceding the 3′-most coding exon of the upstream gene, and the first exon after the 5′-most coding exon of the downstream gene. To confirm EST-supported chimeric transcripts, we designed primers in the 5′-most coding exon of the upstream gene and the 3′-most coding exon of the downstream gene overlapping the EST alignment.
Data availability
All primary data sets used in this study, as well as processed data sets, including computational predictions, the oligonucleotide primers, and the results of experimental verification are available at http://genome.imim.es/datasets/chimeras2005/.
Acknowledgments
We thank Montserrat Corominas and Keith Bradnam for critical reading and help with the manuscript. We also thank the anonymous referees for their very constructive criticisms. N.D. was recipient of a Fulbright fellowship from the U.S Congress and State Department. R.G. acknowledges Plan National de I+D (Spain), the NHGRI ENCODE project, and the European ASD and BIOSAPIENS projects for financial support. S.E.A. acknowledges the NHGRI ENCODE project, Swiss National Science, Foundation, and NCCR Frontiers in Genetics for financial support. E.T.D. is supported by funds from the Wellcome Trust.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4145906.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Akiva, P., Toporik, A., Edelheit, S., Peretz, Y., Diber, A., Shemesh, R., Novik, A., and Sorek, R. 2006. Transcription-mediated gene fusion in the human genome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Athanasiadis, A., Rich, A., and Maas, S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2 e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing, E. and Gautheret, D. 2001. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 11 1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco, E., Parra, G., and Guigó, R. 2003. Using geneid to identify genes. In Current protocols in bioinformatics (eds. A. Baxevanis and D. Davison), Vol. 1, Unit 4.3. John Wiley, New York. [DOI] [PubMed]
- Blow, M., Futreal, P.A., Wooster, R., and Stratton, M.R. 2004. A survey of RNA editing in human brain. Genome Res. 14 2379-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo, R., Reymond, A., Wyss, C., Camara, F., Parra, G., Antonarakis, S.E., Guigó, R., and Eyras, E. 2005. Comparative gene finding in chicken indicates that we are closing in on the set of multi-exonic widely expressed human genes. Nucleic Acids Res. 33 1935-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudevilla, C., Serra, D., Miliar, A., Codony, C., Asins, G., Bach, M., and Hegardt, F.G. 1998. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl. Acad. Sci. 95 12185-12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308 1149-1154. [DOI] [PubMed] [Google Scholar]
- Communi, D., Suarez-Huerta, N., Dussossoy, D., Savi, P., and Boeynaems, J.M. 2001. Cotranscription and intergenic splicing of human P2Y11 and SSF1 genes. J. Biol. Chem. 276 16561-16566. [DOI] [PubMed] [Google Scholar]
- Cox, P.R., Siddique, T., and Zoghbi, H.Y. 2001. Genomic organization of Tropomodulins 2 and 4 and unusual intergenic and intraexonic splicing of YL-1 and Tropomodulin 4. BMC Genomics 2 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420 578-582. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306 636-640. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., Iliopoulos, I., Kyrpides, N.C., and Ouzounis, C.A. 1999. Protein interaction maps for complete genomes based on gene fusion events. Nature 402 86-90. [DOI] [PubMed] [Google Scholar]
- Guigó, R., Dermitzakis, E.T., Agarwal, P., Ponting, C.P., Parra, G., Reymond, A., Abril, J.F., Keibler, E., Lyle, R., Ucla, C., et al. 2003. Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc. Natl. Acad. Sci. 100 1140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, S., Suzuki, Y., Kasai, Y., Morohoshi, K., Yamada, T., Sese, J., Morishita, S., Sugano, S., and Matsushima, K. 2004. 5′-End SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 22 1146-1149. [DOI] [PubMed] [Google Scholar]
- The International Human Genome Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431 931-945. [DOI] [PubMed] [Google Scholar]
- Janin, J., Dumas, C., Morera, S., Xu, Y., Meyer, P., Chiadmi, M., and Cherfils, J. 2000. Three-dimensional structure of nucleoside diphosphate kinase. J. Bioenerg. Biomembr. 32 215-225. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., and Shoemaker, D.D. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302 2141-2144. [DOI] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296 916-919. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389 251-260. [DOI] [PubMed] [Google Scholar]
- Magrangeas, F., Pitiot, G., Dubois, S., Bragado-Nilsson, E., Cherel, M., Jobert, S., Lebeau, B., Boisteau, O., Lethe, B., Mallet, J., et al. 1998. Cotranscription and intergenic splicing of human galactose-1-phosphate uridylyltransferase and interleukin-11 receptor α-chain genes generate a fusion mRNA in normal cells. Implication for the production of multidomain proteins during evolution. J. Biol. Chem. 273 16005-16010. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Ng, H.L., Rice, D.W., Yeates, T.O., and Eisenberg, D. 1999. Detecting protein function and protein–protein interactions from genome sequences. Science 285 751-753. [DOI] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30 13-19. [DOI] [PubMed] [Google Scholar]
- Parra, G., Blanco, E., and Guigó, R. 2000. GeneID in Drosophila. Genome Res. 10 511-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson, J.R. and Fried, V.A. 1992. MacroH2A, a core histone containing a large nonhistone region. Science 257 1398-1400. [DOI] [PubMed] [Google Scholar]
- Poulin, F., Brueschke, A., and Sonenberg, N. 2003. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. J. Biol. Chem. 278 52290-52297. [DOI] [PubMed] [Google Scholar]
- Pradet-Balade, B., Medema, J.P., Lopez-Fraga, M., Lozano, J.C., Kolfschoten, G.M., Picard, A., Martinez, A.C., Garcia-Sanz, J.A., and Hahne, M. 2002. An endogenous hybrid mRNA encodes TWE-PRIL, a functional cell surface TWEAK-APRIL fusion protein. EMBO J. 21 5711-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, A., Camargo, A.A., Deutsch, S., Stevenson, B.J., Parmigiani, R.B., Ucla, C., Bettoni, F., Rossier, C., Lyle, R., Guipponi, M., et al. 2002a. Nineteen additional unpredicted transcripts from human chromosome 21. Genomics 79 824-832. [DOI] [PubMed] [Google Scholar]
- Reymond, A., Marigo, V., Yaylaoglu, M.B., Leoni, A., Ucla, C., Scamuffa, N., Caccioppoli, C., Dermitzakis, E.T., Lyle, R., Banfi, S., et al. 2002b. Human chromosome 21 gene expression atlas in the mouse. Nature 420 582-586. [DOI] [PubMed] [Google Scholar]
- Roginski, R.S., Mohan Raj, B.K., Birditt, B., and Rowen, L. 2004. The human GRINL1A gene defines a complex transcription unit, an unusual form of gene organization in eukaryotes. Genomics 84 265-276. [DOI] [PubMed] [Google Scholar]
- Sadek, C.M., Damdimopoulos, A.E., Pelto-Huikko, M., Gustafsson, J.A., Spyrou, G., and Miranda-Vizuete, A. 2001. Sptrx-2, a fusion protein composed of one thioredoxin and three tandemly repeated NDP-kinase domains is expressed in human testis germ cells. Genes Cells 6 1077-1090. [DOI] [PubMed] [Google Scholar]
- Snel, B., Bork, P., and Huynen, M. 2000. Genome evolution. Gene fusion versus gene fission. Trends Genet. 16 9-11. [DOI] [PubMed] [Google Scholar]
- Takahara, T., Tasic, B., Maniatis, T., Akanuma, H., and Yanagisawa, S. 2005. Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol. Cell 18 245-251. [DOI] [PubMed] [Google Scholar]
- Thomson, T.M., Lozano, J.J., Loukili, N., Carrio, R., Serras, F., Cormand, B., Valeri, M., Diaz, V.M., Abril, J., Burset, M., et al. 2000. Fusion of the human gene for the polyubiquitination coeffector UEV1 with Kua, a newly identified gene. Genome Res. 10 1743-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13 308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.F., Hartman, S.J., Schroeder, D.I., Otillar, R.P., and Myers, R.M. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14 62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420 520-562. [DOI] [PubMed] [Google Scholar]
- Zaphiropoulos, P.G. 1999. RNA molecules containing exons originating from different members of the cytochrome P450 2C gene subfamily (CYP2C) in human epidermis and liver. Nucleic Acids Res. 27 2585-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://bind.ca/; BIND databases.
- http://genome.cse.ucsc.edu/; UCSC genome browser database.
- http://genome.imim.es/datasets/chimeras2005/; Supplemental materials and results.
- http://genome.ucsc.edu/ENCODE/regions.html; UCSC ENCODE repository.
- http://string.embl.de/; STRING server.
- http://vega.sanger.ac.uk/; Vertebrate Annotation Database (VEGA).
- http://www.ncbi.nlm.nih.gov/RefSeq/; NCBI Reference Sequence (RefSeq).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All primary data sets used in this study, as well as processed data sets, including computational predictions, the oligonucleotide primers, and the results of experimental verification are available at http://genome.imim.es/datasets/chimeras2005/.