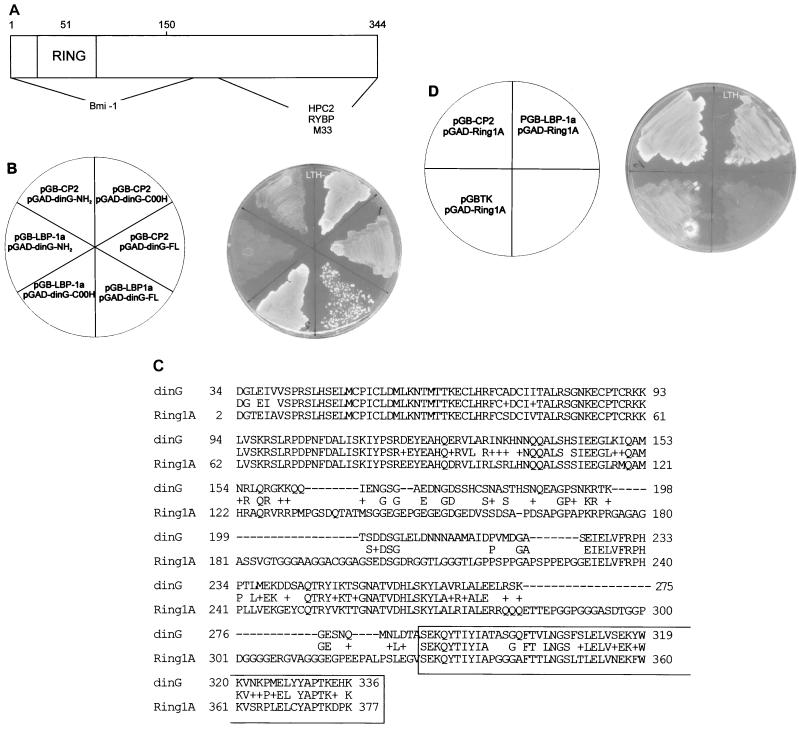
FIG. 3.
The RING domain of dinG is not required for the interaction with CP2/LBP-1a. (A) Schematic representation of the dinG protein. The RING finger domain is marked, and the regions interacting with Bmi-1 and HPC2 are indicated. The region of the murine homologue of dinG, Ring1B, that interacts with RYBP and M33 is also shown. (B) Mapping the CP2/LBP-1a interaction domain of dinG. (Left) The S. cerevisiae reporter strain HF7C was transformed with the indicated plasmids. pGB-CP2-260 and pGB-LBP-1a are detailed in the legend to Fig. 1. pGAD-dinG-FL contains the entire dinG cDNA fused to the GAL4AD. pGAD-dinG-NH2 contains a dinG cDNA fragment encoding amino acids 1 to 165 fused to GAL4AD, and pGAD-dinG-COOH contains a dinG cDNA fragment encoding amino acids 286 to 336 fused to GAL4AD. (Right) Transformants were streaked onto synthetic medium plates lacking tryptophan, leucine, and histidine (LTH−) and incubated at 30°C for 3 days. (C) Sequence comparison of dinG and Ring1A. The deduced amino acid sequences encoded by dinG and Ring1A cDNAs are aligned using the BLAST algorithm (2). Amino acid identity is shown, and similarities are indicated by (+). Dashes represent gaps introduced to maximize the alignment. The region of dinG that interacts with CP2/LBP-1a is boxed. (D) Ring1A interacts with CP2/LBP-1a. (Left) The S. cerevisiae reporter strain HF7C was transformed with the indicated plasmids. pGB-CP2-260 and pGB-LBP-1a are detailed in the legend to Fig. 1. pGBTK has been described previously. pGAD-Ring1A contains a Ring1A cDNA fragment encoding amino acids 327 to 377 fused to GAL4AD. (Right) Transformants were streaked onto synthetic medium plates lacking tryptophan, leucine, and histidine (LTH−) and incubated at 30°C for 3 days.