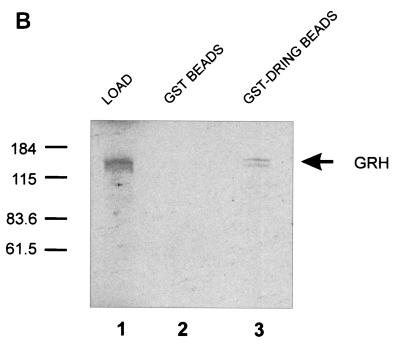
FIG. 6.
DRING, the Drosophila homologue of dinG, interacts with GRH. (A) Sequence comparison of dinG and DRING. The deduced amino acid sequences encoded by dinG and dring cDNAs were aligned by using the BLAST algorithm (2). Amino acid identity is shown, and similarities are indicated by plus signs. Dashes represent gaps introduced to maximize the alignment. The region of dinG that interacts with CP2/LBP-1a is boxed. (B) GST chromatography of DRING and GRH. A fusion protein between GST and DRING (GST-DRING) or GST alone was expressed in E. coli, and 1 mg of protein was bound to glutathione-Sepharose beads. The beads were then incubated with 2 ml of 35S-labeled in vitro-translated GRH in binding buffer for 1 h at room temperature (see Materials and Methods). After extensive washing, the beads were resuspended in SDS loading buffer and subjected to SDS-PAGE. The migrations of the GRH load and the molecular mass standards (in kilodaltons) are indicated.