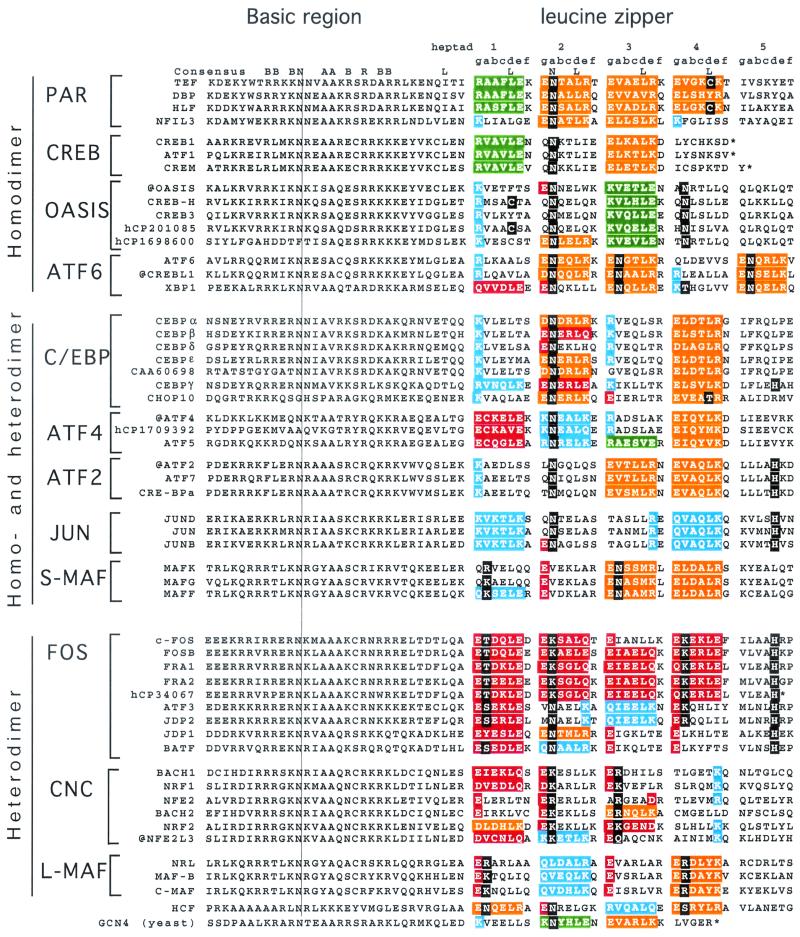
FIG. 3.
Amino acid sequence of all 53 human B-ZIP domains. Proteins are placed in to 12 groups based on predicted dimerization properties. We have placed the well-studied yeast B-ZIP protein GCN4 at the bottom of the figure for reference. The natural C terminus is denoted with an asterisk. Leucine zipper heptads are grouped (gabcdef) to help visualize potential g↔e′ pairs. If both the g and e positions contain charged amino acids, we have colored the gabcde amino acids. Our description of “attractive” and “repulsive” refers to pairs that would be present in the homodimer. We use the four colors to describe the g↔e′ pairs. Green is for the attractive basic-acidic pairs (R↔E and K↔E), orange is for the attractive acidic-basic pairs (E↔R, E↔K, D↔R, and D↔K), red is for the repulsive acidic pairs (E↔E, E↔D, E↔Q, and Q↔E), and blue is for the repulsive basic pairs (K↔K, R↔K, Q↔K, R↔Q, and K↔Q (Table 3). If only one of the two amino acids in the g↔e′ pair is charged, we color only that amino acid: red if it is acidic and blue if it is basic. If the a or d positions contain polar or charged amino acids, they are colored black.