Abstract
Pleckstrin homology (PH) domains play diverse roles in cytoskeletal dynamics and signal transduction. Split PH domains represent a unique subclass of PH domains that have been implicated in interactions with complementary partial PH domains ‘hidden' in many proteins. Whether partial PH domains exist as independent structural units alone and whether two halves of a split PH domain can fold together to form an intact PH domain are not known. Here, we solved the structure of the PHN–PDZ–PHC tandem of α-syntrophin. The split PH domain of α-syntrophin adopts a canonical PH domain fold. The isolated partial PH domains of α-syntrophin, although completely unfolded, remain soluble in solution. Mixing of the two isolated domains induces de novo folding and yields a stable PH domain. Our results demonstrate that two complementary partial PH domains are capable of binding to each other to form an intact PH domain. We further showed that the PHN–PDZ–PHC tandem forms a functionally distinct supramodule, in which the split PH domain and the PDZ domain function synergistically in binding to inositol phospholipids.
Keywords: lipid binding, PDZ domain, split PH domain, supramodule, syntrophin
Introduction
Syntrophins are a family of multi-domain scaffold proteins that associate with dystrophin and the dystrophin-related proteins, utrophin and dystrobrevin (Adams et al, 1993; Ahn et al, 1994; Yang et al, 1994). All five members (α, β1, β2, γ1, and γ2) of the syntrophin family share the same domain organization: an N-terminal split PH domain containing an embedded PDZ domain, a central PH domain, and a C-terminal syntrophin unique domain (SU) (Figure 1A; Adams et al, 1993; Ahn et al, 1994; Yang et al, 1994; Piluso et al, 2000). The second PH domain and the SU domain of syntrophins are responsible for binding to dystrophin, utrophin, and dystrobrevin (Kramarcy et al, 1994; Ahn and Kunkel, 1995; Suzuki et al, 1995; Kachinsky et al, 1999). A large number of signaling proteins that bind to the PDZ domain of syntrophins have been identified. These proteins include neuronal nitric oxide synthase (nNOS) (Brenman et al, 1996; Adams et al, 2001), aquaporin-4 (Adams et al, 2001; Neely et al, 2001), voltage-gated sodium channels (Gee et al, 1998; Ou et al, 2003), potassium channels (Connors et al, 2004; Leonoudakis et al, 2004), serine/threonine protein kinases (Hasegawa et al, 1999; Lumeng et al, 1999), the ATP-binding cassette transporter A1 (Buechler et al, 2002), and others. The first PH domain of syntrophin has been implicated in phospholipid binding (Chockalingam et al, 1999). Since none of the defined domains contain intrinsic enzyme activity, syntrophins are regarded as adaptors that function by targeting PDZ-binding partners to the membrane via the dystrophin complex (Albrecht and Froehner, 2002).
Figure 1.
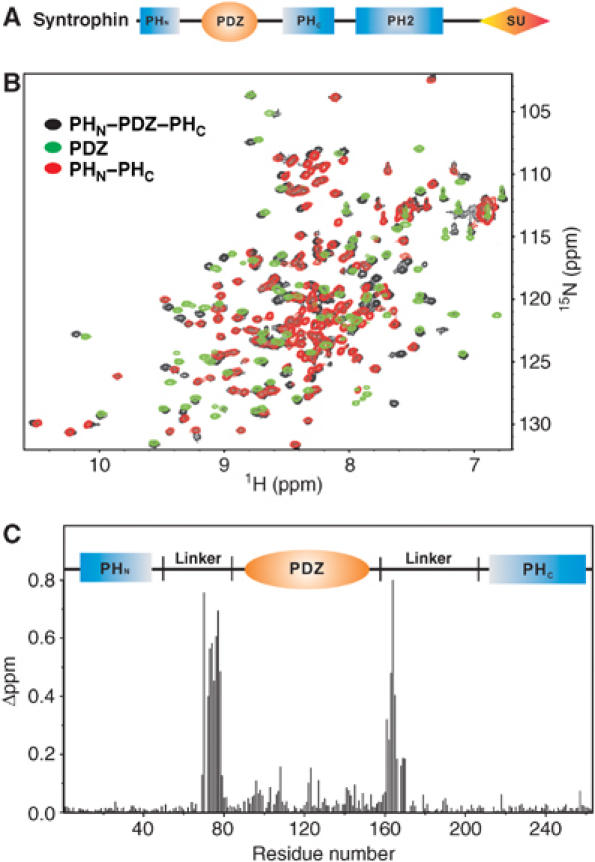
Comparison of the split PH and PDZ domains in the PHN–PDZ–PHC tandem and in their respective isolated state. (A) Schematic diagram showing the domain organization of syntrophins. (B) Superposition plots of 1H, 15N HSQC spectra of the PHN–PDZ–PHC tandem (black), the isolated PDZ domain (green), and the joint PHN–PHC domain (red). (C) Plot of chemical shift changes as a function of residue number of the split PH and PDZ domains in the PHN–PDZ–PHC tandem and in their respective isolated forms. The combined 1H and 15N chemical shift changes are defined as:  ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts between the PHN–PDZ–PHC tandem and the isolated PH and PDZ domains, respectively. The scaling factor (αN) used to normalize the 1H and 15N chemical shifts is 0.17. The domain organization of the PHN–PDZ–PHC tandem is indicated at the top of the plot.
ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts between the PHN–PDZ–PHC tandem and the isolated PH and PDZ domains, respectively. The scaling factor (αN) used to normalize the 1H and 15N chemical shifts is 0.17. The domain organization of the PHN–PDZ–PHC tandem is indicated at the top of the plot.
PH domains are abundant protein modules that play critical roles in cellular signaling and cytoskeletal organization (Lemmon and Ferguson, 2000). Despite their low amino-acid sequence identity, all PH domains with known structures contain a conserved core structure composed of a partially open, two-sheeted β-barrel with one end of the barrel capped with a C-terminal α-helix (Ferguson et al, 1994; Macias et al, 1994; Yoon et al, 1994; Fushman et al, 1995; Lemmon and Ferguson, 2000). The first β-sheet is composed of four antiparallel β-strands (β1–β4), and the second β-sheet contains three strands (β5–β7). The best characterized function of PH domains is binding to inositol phospholipids (Lemmon and Ferguson, 2000). Only a minority of PH domains (e.g., the PH domains from phospholipase Cδ, Bruton's tyrosine kinase, and protein kinase B; Fukuda et al, 1996; Frech et al, 1997; Rameh et al, 1997; Kavran et al, 1998; Yu et al, 2004) are capable of binding to lipids with high affinity and specificity. Some PH domains are known to be weak, nonspecific membrane phosphoinositide binders (Yu et al, 2004), while others interact with proteins (e.g., the PH domain of β-adrenergic receptor; Touhara et al, 1994). However, the function of the majority of PH domains is unknown (Yu et al, 2004).
The PH domain organization of syntrophins is unique. Based on the amino-acid sequence analysis, the N-terminal PH domain is split into two halves by insertion of the PDZ domain (Figure 1A). The insertion of the PDZ domain in the middle of the N-terminal PH domain is a highly conserved molecular characteristic of all isoforms of syntrophins, suggesting the potential functional significance of this arrangement. However, it is not known whether the split PH domains of syntrophins can fold properly into a canonical PH domain structure. Additionally, the functional significance (if any) of the PDZ domain-mediated PH domain splitting has not been addressed. Another example of PH domain splitting by insertion of other protein–protein interaction domains is phospholipase Cγ1. The C-terminal PH domain of phospholipase Cγ1 contains an insert of a tandem array of SH2–SH2–SH3 domains (Chang et al, 2002). It was reported that the split halves of the phospholipase Cγ1 PH domain may function independently in binding to target proteins (Chang et al, 2002, 2005). Very recently, van Rossum et al (2005) reported that the C-terminal half of the phospholipase Cγ1 PH domain (PLCγ1–PHC) can interact with a short fragment of peptide sequence in the TRPC3 ion channel. The authors suggested that the PLCγ1–PHC-binding segment of TRPC3 represents a complementary partial PH domain ‘hidden' in the ion channel. Binding of the two partial PH domain fragments from PLCγ1 and TRPC3, respectively, forms a functional PH domain capable of binding to specific lipids and regulating surface expression of the TRPC3 ion channel (van Rossum et al, 2005). Given the potentially wide distribution of split PH domains in diverse proteins and enzymes, the work presented by van Rossum et al (2005) suggests a novel mode of function of many PH domains. To advance this important hypothesis, it is critical to know whether the complex formed by two fragments from PLCγ1 and TRPC3 (or PLCγ1 and translational elongation factor 1α (Chang et al, 2002)) or from other proteins with split PH domains can indeed assume a PH domain-like fold.
In this work, we solved the three-dimensional structure of the PHN–PDZ–PHC supramodule of α1-sytrophin by NMR spectroscopy. We show, for the first time, that the two halves of the split PH domain associate with each other, forming a canonical, intramolecular PH domain fold. The insertion of the PDZ domain has negligible impact on the conformation of the PH domain. We found, however, that the split PHN–PDZ–PHC supramodule has distinct phosphoinositide lipid-binding properties when compared to an isolated PH domain formed by simply connecting the PHN and the PHC with a linker sequence or the PDZ domain alone.
Results
The split PH domain of the PHN–PDZ–PHC tandem from α-syntrophin adopts the same fold as the two halves linked together
We chose the PHN–PDZ–PHC tandem of α-syntrophin for detailed structural analysis as its biological function is best known within the syntrophin family (Kameya et al, 1999; Adams et al, 2000, 2001; Neely et al, 2001; Hosaka et al, 2002). The recombinant PHN–PDZ–PHC tandem of α-syntrophin (residues 2–264, referred to hereafter as PHN–PDZ–PHC) was eluted at molecular mass indicative of a stable monomer when analyzed by analytical gel filtration chromatography (data not shown). The well-dispersed 1H, 15N HSQC spectrum of PHN–PDZ–PHC indicates that both the split PH domain and the PDZ domain are folded (Figure 1B, black peaks). Next, we deleted the PDZ domain from the PHN–PDZ–PHC, and purified a covalently linked form of PH domain composed of PHN–PHC. Analytical gel filtration analysis of the joined PHN–PHC showed that the protein also exists as a stable monomer in solution (data not shown). The 1H, 15N HSQC spectrum of PHN–PHC (Figure 1B and C, red peaks) overlaps very well with a subset of peaks which originate from the two split halves of the PH domain in PHN–PDZ–PHC. The HSQC peaks of the isolated PDZ domain (green) overlap less well with the peaks from the PDZ domain in the PHN–PDZ–PHC tandem (Figure 1B). After assigning the chemical shifts of the PHN–PDZ–PHC tandem as well as the isolated PH and PDZ domains, we plotted the chemical shift differences of backbone amides as a function of residue number between the PHN–PDZ–PHC tandem and the isolated PH and PDZ domains (Figure 1C). The largest chemical shift differences are located in the linker regions that connect the two halves of the PH domain with the PDZ domain. Such large chemical shift changes are expected when the PDZ domain is removed from the PHN–PDZ–PHC tandem. The two halves of the PH domain showed minimal shift differences whether they are in the PHN–PDZ–PHC tandem form or in the joined PHN–PHC form, indicating that the two halves of the PH domain of α-syntrophin can fold into the same stable structure with or without the PDZ insertion. The residues that show small but significant differences in the two forms of PDZ domain are concentrated within an area in the vicinity of the two termini of the domain (Supplementary Figure 1). Taken together, our NMR and gel filtration chromatography studies demonstrate that the split PH domain of α-syntrophin can adopt a stable, intramolecular structure.
Structure of the PHN–PDZ–PHC supramodule
To determine if the split PH domain of α-syntrophin folds into a canonical PH domain structure, we solved the three dimensional structures of the PHN–PDZ–PHC tandem as well as the joined PHN–PHC domain and the isolated PDZ domain by NMR spectroscopy (Figure 2 and Table I, Supplementary Figures 2 and 3, and Supplementary Table 1). Consistent with the data shown in Figure 1, the PHN–PDZ–PHC tandem contains two separate domains. The PHN and PHC fragments fold together to form a canonical PH domain structure containing seven β-strands and one C-terminal α-helix (Figure 2A–C). The PHN half is composed of three β-strands (β1–β3), and the PHC half contains the remaining four β-strands (β4–β7) and the C-terminal α-helix. Inserted at the β3/β4-loop of the PH domain is a well-folded PDZ domain and two long linkers (Figure 2C–E). No interdomain NOEs between the split PH domain and the PDZ domain were observed; therefore, the orientations of the two domains were not defined. Potential long-range interactions between the split PH domain and the PDZ domain were further investigated by NMR-based approaches. First, we measured the residual dipolar coupling constants of the backbone amides of the PHN–PDZ–PHC tandem by aligning the protein in a 4.5% positively charged copolymer containing 50% of (3-acrylamidopropyl)-trimethylammonium chloride and 50% of acrylamide (Cierpicki and Bushweller, 2004). The alignment tensors of the split PH domain and the PDZ domain in this alignment medium were very different (data not shown), indicating that the orientations of the two domains are not fixed in the PHN–PDZ–PHC tandem. We further probed the orientation of the split PH domain and the PDZ domain in the PHN–PDZ–PHC tandem by tagging the split PH domain with S-cysteaminyl-EDTA, an EDTA derivative used to detect long-range interactions in proteins using paramagnetic pseudo-contact shifts (Ikegami et al, 2004; Pintacuda et al, 2004). Addition of Co2+ to the S-cysteaminyl-EDTA-tagged PHN–PDZ–PHC induced chemical shift changes limited to the split PH domain (Supplementary Figure 4). This paramagnetic pseudo-contact shift perturbation data further indicated that the orientation of the split PH domain and the PDZ domain in the PHN–PDZ–PHC tandem is not fixed.
Figure 2.
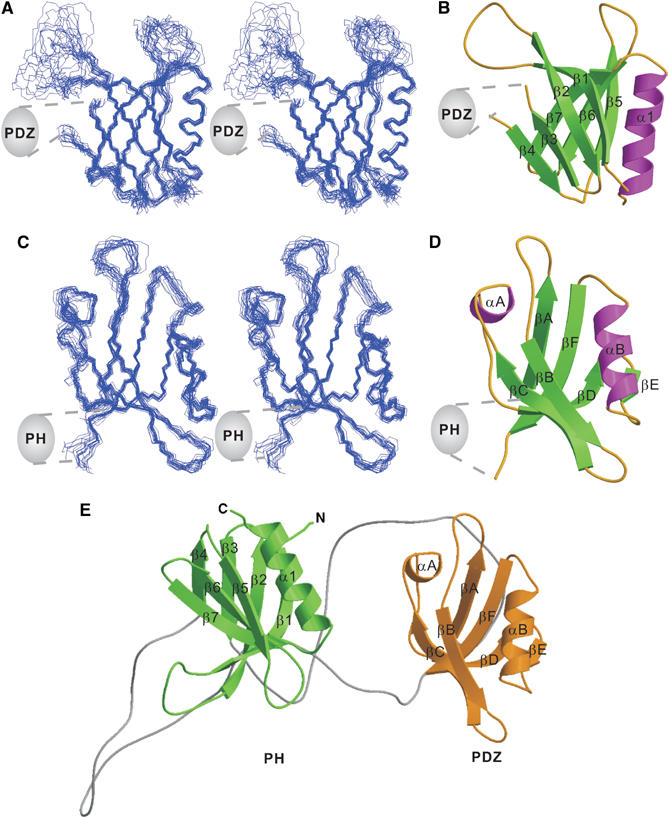
Structure of the PHN–PDZ–PHC tandem. (A) Stereo view showing the backbones of 20 superimposed NMR-derived structures of the split PH domain from the PHN–PDZ–PHC tandem. Since the orientation between the split PH domain and the PDZ domain in the PHN–PDZ–PHC tandem is not fixed, we chose to present the structures of each domain separately. (B) Ribbon diagram of a representative NMR structure of the split PH domain. The insertion of the PDZ domain in the β3/β4-loop of the split PH domain is indicated. (C) Stereo view plot of 20 superimposed NMR structures of the PDZ domain from the PHN–PDZ–PHC tandem. (D) Ribbon diagram drawing of the PDZ domain structure. The split PH domain connected to the βA and βF of the domain is indicated. (E) Ribbon diagram drawing of a representative NMR structure of the PHN–PDZ–PHC tandem. In this drawing, the split PH domain is in green, and the PDZ domain is in gold.
Table 1.
Structural statistics for the final 20 structures of the PHN–PDZ–PHC tandema
| Distance restraints | |
|---|---|
| Intraresidue (i−j=0) | 1160 |
| Sequential (∣i−j∣=1) | 627 |
| Medium range (2⩽∣i−j∣⩽4) | 206 |
| Long range (∣i−j∣>5) | 607 |
| Hydrogen bonds | 140 |
| Total | 2740 |
| Dihedral angle restraints | |
| Φ | 94 |
| Ψ | 90 |
| Total | 184 |
| Mean r.m.s. deviations from the experimental restraints | |
| Distance (Å) | 0.012±0.000 |
| Dihedral angle (deg) | 0.102±0.015 |
| Mean r.m.s. deviations from idealized covalent geometry | |
| Bond (Å) | 0.002±0.000 |
| Angle (deg) | 0.306±0.007 |
| Improper (deg) | 0.149±0.010 |
| Mean energies (kcal mol−1) | |
| ENOEb | 27.75±0.71 |
| Ecdihb | 0.12±0.03 |
| EL–J | −488.03±31.34 |
| Ramachandran plotc | |
| (Residues 2–45, 79–163, 205–264) residues in the most favorable regions | 69.3 |
| Additional allowed regions | 26.1 |
| Generously allowed regions | 3.0 |
| Disallowed regions | 1.6 |
| Atomic r.m.s. difference (Å)d (residues 9–15, 29–44, 206–219, 229–262 in PH) | |
| Backbone heavy atoms (N, Cα, and C′) | 0.54 |
| Heavy atoms (residues 80–85, 93–162 in PDZ) | 1.14 |
| Backbone heavy atoms (N, Cα, and C′) | 0.51 |
| Heavy atoms |
1.09 |
| aNone of the structures exhibits distance violations greater than 0.3 Å or dihedral angle violations greater than 4°. | |
| bThe final values of the square-well NOE and dihedral angle potentials were calculated with force constants of 50 kcal mol−1 Å−2 and 200 kcal mol−1 rad−2, respectively. | |
| cThe program Procheck (Laskowski et al, 1996) was used to assess the overall quality of the structures. | |
| dThe precision of the atomic coordinates is defined as the average r.m.s. difference between 20 final structures and the mean coordinates of the protein. | |
Consistent with the nearly complete overlap of the peaks in the NMR spectra (Figure 1B), the structure of the joined PHN–PHC is essentially the same as the split PH domain structure in the PHN–PDZ–PHC tandem (a root-mean-square difference of 1.0 Å for the backbones of the two structures; Supplementary Figure 2). The structure of the isolated PDZ domain is also essentially the same as the domain in the PHN–PDZ–PHC tandem (Supplementary Figure 3; also see Schultz et al, 1998a).
Since split PH domains have been observed in several proteins in addition to syntrophins, it is critical to understand whether the inserted sequences between the two halves of a split PH are required for the domain to fold. This issue becomes particularly relevant as the C-terminal half of the PLCγ1 split PH domain (PLCγ1–PHC) was suggested to bind to a partial PH fragment hidden in TRPC3 instead of the PHN within PLCγ1 (van Rossum et al, 2005). To address this question, we replaced the PDZ domain of PHN–PDZ–PHC with an eight-residue peptide fragment that can be cleaved by protease 3C (referred to as PHN–‘C'–PHC; Figure 3A). The NMR spectra indicated that the purified PHN–‘C'–PHC folds into a structure indistinguishable from that of the joined PHN–PHC domain (Figures 1B and 3B). Digestion of PHN–‘C'–PHC with protease 3C produces two fragments with molecular masses corresponding to PHN and PHC, respectively (Figure 3A). The NMR spectrum of the protease 3C-cleaved PHN–‘C'–PHC is essentially identical to that of the uncleaved protein (Figure 3B), indicating that the covalent linkage between PHN and PHC (and hence the PDZ domain) is dispensable for the folding of the split PH domain in syntrophins.
Figure 3.
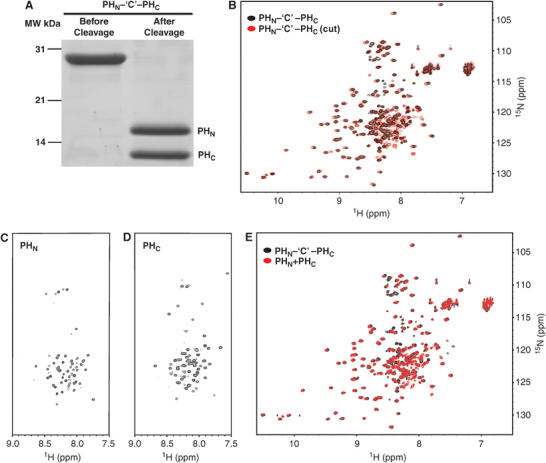
Folding and interaction of the two isolated, partial PH domain fragments. (A) SDS–PAGE showing the purification of the joined PHN–‘C'–PHC domain and cleavage of the domain into PHN and PHC fragments. (B) Overlay plot of the 1H, 15N HSQC spectra of the joined PHN–‘C'–PHC (black) and its protease cleaved form (red). 1H, 15N HSQC spectra of the isolated PHN (C) and PHC (D) fragments. (E) Overlay plot of the 1H, 15N HSQC spectra of the joined PHN–‘C'–PHC (black) and the 1:1 mixture of the two halves of the split PH domain (red).
To investigate whether two partial but complementary PH domains can indeed interact with each other intermolecularly, we studied the structure of the two isolated PHN and PHC fragments of α-syntrophin, as well as a 1:1 mixture of the two fragments. The cleaved His6-tagged PHN was separated from PHC under denatured condition by passing the protease-cleaved mixture through a Ni2+-NTA affinity column. The denaturant (urea) was subsequently dialyzed away from each half of the purified PH domain (Figure 3A). The NMR spectra of the two isolated fragments showed that each domain alone is completely unfolded, but remains soluble and stable in solution, indicating that partial PH domains can exist as independent structural units (Figure 3C and D). When mixed together, the interaction of the two partial PH domains induces de novo folding of both fragments. Further, the NMR data showed that the two partial PH domains fold into an intact PH domain structure indistinguishable from that of the split PH domain in the PHN–PDZ–PHC tandem (Figure 3E). The PH domain formed by the two isolated halves remains stably folded for a number of days under our NMR measurement conditions.
The α-syntrophin PHN–PDZ–PHC tandem functions as a supramodule with distinct inositol phospholipid-binding properties
Since the split PH domain of PHN–PDZ–PHC can fold into a canonical PH domain structure, we assayed the binding of the PHN–PDZ–PHC tandem to liposomes prepared from total bovine brain lipids (Figure 4). The PHN–PDZ–PHC tandem was found to bind efficiently to these liposomes (Figure 4B, top panel). The interaction between the PHN–PDZ–PHC tandem and the brain liposome is dose-dependent, with an estimated Kd value of ∼5 μM (Figure 4C). To our surprise, binding of the joined PHN–PHC domain to liposomes is much weaker than the PHN–PDZ–PHC tandem, even though the PHN–PHC domain folds into the same structure as the split PH domain in the PHN–PDZ–PHC tandem (Figure 4B, second panel). Since PDZ domains have been shown to bind directly to inositol phospholipids (Zimmermann et al, 2002; Mortier et al, 2005), we tested whether the PDZ domain of α-syntrophin could also bind to lipids, using the same brain liposome-binding assay. The α-syntrophin PDZ domain displayed some binding to lipids, albeit with a much weaker binding capacity when compared to the PHN–PDZ–PHC tandem (Figure 4B, third panel). Further, the enhanced lipid-binding avidity of α-syntrophin requires that the split PH domain and the PDZ domain be covalently linked, as a mixture containing the isolated PHN–PHC domain and the PDZ domain binds to brain liposomes only weakly (Figure 4B, bottom panel). To ensure that the altered lipid-binding properties of the mutants shown in Figure 4 did not result from mutation-induced protein stability changes, we compared the stabilities of these mutants (PHN–PHC and isolated PDZ domain) with that of the wild-type PHN–PDZ–PHC tandem. Both the temperature- and urea-dependent denaturation profiles of the mutants and the PHN–PDZ–PHC tandem are highly similar (Supplementary Figure 5), indicating that the decreased lipid-binding avidities of the mutants are not the result of their compromised stability.
Figure 4.
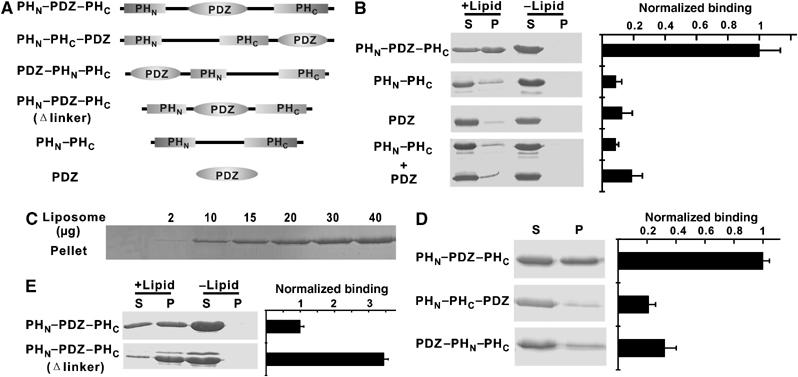
The PHN–PDZ–PHC supramodule binds to brain liposomes with enhanced avidity. (A) Schematic diagrams showing the various forms of proteins purified for liposome-binding assays. (B) Brain liposome-binding assays of the PHN–PDZ–PHC tandem and its fragments. ‘S' and ‘P' denote proteins recovered in the supernatants and pellets, respectively, in the centrifugation-based liposome-binding assays. (C) Dose-dependent binding between the PHN–PDZ–PHC tandem and the brain liposome. In this assay, the amount of the PHN–PDZ–PHC tandem is fixed at 10 μM, and the concentration of liposome varies. The liposome-bound PHN–PDZ–PHC tandem is recovered in the pellet in the binding assay. (D) Lipid binding of the two mutants of the PHN–PDZ–PHC tandem. In these two mutants, the PDZ domain was placed either at the front or after the split PH domain. In these mutants, the PH and the PDZ domains were connected by a flexible ‘Gly–Ser–Gly–Gly–Ser–Gly–Gly–Ser–Gly–Ser' linker. (E) Lipid binding of a PHN–PDZ–PHC tandem mutant with the two connecting linkers shortened to six residues. Amino-terminal His6-tagged proteins were used in all of the brain liposome-binding assays.
It is conceptually easy to rationalize that, when covalently connected, two relatively weak lipid-binding domains can function synergistically to increase the lipid-binding avidity of the PHN–PDZ–PHC tandem. However, it is intriguing that the PH domain of every member of the syntrophin family contains a PDZ domain inserted in the middle of its primary sequence; yet, the folding and structure of the PH domains are not affected by this insertion. We hypothesized that the position-specific insertion of the weak lipid-binding PDZ domain in the middle of the PH domain may somehow specifically modify the lipid-binding properties of the PHN–PDZ–PHC tandem. To test this hypothesis, we created two mutants of the PHN–PDZ–PHC tandem by placing the PDZ domain either in front of or after the joined PHN–PHC domain. To avoid artificial conformational restraints, a 10-residue flexible linker (‘Gly–Ser–Gly–Gly–Ser–Gly–Gly–Ser–Gly–Ser') was inserted between the PDZ domain and the joined PHN–PHC domain in both mutants. Both mutants displayed weaker lipid binding compared to the PHN–PDZ–PHC tandem (Figure 4D). In the protein stability test, both PDZ–PHN–PHC and PHN–PHC–PDZ showed similar stability to the PHN–PDZ–PHC tandem (Supplementary Figure 5). Our data indicate that splitting the PH domain by insertion of the PDZ domain creates a PHN–PDZ–PHC supramodule with unique lipid-binding properties.
If the linkers between the split PH domain and the PDZ domain in the PHN–PDZ–PHC tandem are entirely flexible, we should not observe the obvious differences in lipid-binding avidity between the wild-type PHN–PDZ–PHC and the two mutants shown in Figure 4D. To address this, we analyzed the structural feature of the two linkers connecting the two halves of the split PH domain and the PDZ domain in detail. The secondary structure-induced 13Cα and 13Cβ chemical shifts of the split PH domain in the PHN–PDZ–PHC tandem showed that the two linkers are partially structured (Supplementary Figure 6). Specifically, the linker connecting the PHN and PDZ contains a β-strand-like structure immediately following the β3 of PHN and an α-helix-like segment N-terminal to the PDZ domain. The linker connecting the PHC and PDZ contains two partial β-strands N-terminal to the β4 strand of PHC. Presumably, the two partially structured linkers restrict the degree of motion between the split PH domain and the PDZ domain to a certain extent, thereby promoting binding of the PHN–PDZ–PHC tandem to liposomes. The small but significant chemical shift differences between the isolated PDZ domain and the domain in the PHN–PDZ–PHC tandem (Supplementary Figure 1) are consistent with the notion that the linkers may influence the motion of the PDZ domain. To experimentally test if the interdomain motion between the split PH domain and the PDZ domain is correlated with the lipid-binding avidity of the protein, we shortened both linkers of the PHN–PDZ–PHC tandem to six residues, and found that the resulting mutant binds to liposomes with a significantly higher avidity than the wild-type protein (Figure 4E).
The binding specificity of the PHN–PDZ–PHC tandem towards different lipids was assayed using membrane-immobilized lipid strips. Of the 15 lipids tested, the PHN–PDZ–PHC tandem displayed strong binding to several phosphoinositides including PI(3,5)P2 and PI(5)P, modest binding to PI(3,4)P2 and PI(4,5)P2, and weak binding to other inositol phospholipids (Figure 5A). No binding could be detected between the PHN–PDZ–PHC tandem and two major components of mammalian membrane lipids, phosphatidylcholine and phosphatidylethanolamine, suggesting that the interaction between the PHN–PDZ–PHC tandem and phosphoinositides is relatively specific. However, the stereo-specificity among different phosphoinositides is not very high. Consistent with the data derived from the brain liposome assay, the isolated PHN–PHC domain, the isolated PDZ domain, and the mutants with the PDZ domain position swapped all displayed very low lipid-binding affinity in the lipid strip-binding assay (data not shown). Since lipid strip-based assay is very qualitative, we used a fluorescence perturbation assay to quantitatively measure affinity between the PHN–PDZ–PHC tandem and PI(3,5)P2. Binding was monitored via fluorescence change of a 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) fluorophore-labeled protein (Papayannopoulos et al, 2005) upon addition of vesicles (phosphatidyl choline (PC), phosphatidyl serine (PS), and (PI(3,5)P2) at ratios of 70:20:10 and 60:20:20, respectively). No detectable binding of the PHN–PDZ–PHC tandem is observed to vesicles containing only PC/PS (80:20). Strong binding is observed to vesicles containing 10 and 20% PI(3,5)P2 (apparent Kd ∼5.1 and 3.9 μM, respectively) (Figure 5B), indicating that the interaction between the PHN–PDZ–PHC tandem and phosphoinositides is specific.
Figure 5.
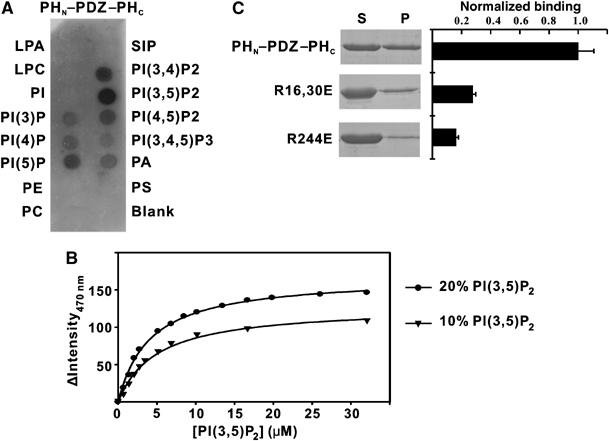
Lipid-binding specificity of the PHN–PDZ–PHC supramodule using phospholipid strip overlay assay. (A) Binding of the PHN–PDZ–PHC tandem to the phospholipids spotted on a cellulose membrane. The amount of lipid per spot was 100 pmol. Abbreviations for the lipids: S1P: sphingosine-1-phosphate; LPA: lysophosphatidic acid; LPC: lysophosphocholine; PE: phosphatidylethanolamine; PS: phosphatidylserine; PA: phosphatidic acid; PC: phosphatidylcholine; PtdIns: phosphatidylinositol. The amount of protein used in the assay is 0.5 μg/ml. (B) Binding isotherms of PHN–PDZ–PHC with PC/PS liposomes containing either 10 or 20% PI(3,5)P2. (C) Identification of the residues in the split PH domain of the PHN–PDZ–PHC tandem that are involved in the lipid binding.
To identify the phosphoinositide-binding site in the PH domain, we mutated several positively charged residues in the β1/β2-loop (Arg16 and Arg30) and in the β7-strand (Arg244) of the split PH domain. These positively charged residues are clustered at one end of the split PH domain (data not shown), and are expected to be involved in lipid binding (Ferguson et al, 1995; Lemmon and Ferguson, 2000; Lietzke et al, 2000; Thomas et al, 2002). Substitutions of Arg16 and Arg30 or Arg244 by Glu significantly reduced the lipid-binding capacity of the PHN–PDZ–PHC tandem, indicating that these positively charged residues are indeed required for lipid binding (Figure 5C). We tried to map the lipid-binding site of the split PH domain by titrating the joined PHN–PHC domain or the PHN–PDZ–PHC tandem (both uniformly labeled with 15N) with water-soluble head groups of phosphoinositides that display significant binding to the PHN–PDZ–PHC tandem. However, we were not able to detect specific interactions between the head groups of these phosphoinositides with the split PH domains. In agreement with a very recent study (Mortier et al, 2005), we were not able to detect binding of the isolated PDZ domain to water-soluble head groups of phosphoinositides.
Next, we studied the potential impact of the PDZ ligands on the lipid binding of the PHN–PDZ–PHC tandem. We found that addition of an excess amount of α-syntrophin PDZ ligand (a synthetic peptide containing a consensus α-syntrophin PDZ-binding motif ‘S–X–V*', or the nNOS PDZ domain) did not significantly affect the lipid-binding property of the PHN–PDZ–PHC tandem. Thus, α-syntrophin PDZ ligands do not interfere with the lipid-binding capacity of the PHN–PDZ–PHC supramodule (Figure 6A and data not shown). Our data also imply that the lipid-binding site and the PDZ ligand-binding site on the α-syntrophin PDZ domain do not overlap with each other. Consistent with this observation, we found that mutations of the two positively charged residues in the carboxyl-binding loop of the target recognition groove of the PDZ domain (Arg85 and Arg86 to Ser) had no impact on lipid binding of the PHN–PDZ–PHC tandem (Figure 6B). In a reverse experiment, we showed that the binding of lipids to the PHN–PDZ–PHC tandem does not affect the binding of α-syntrophin PDZ domain to the nNOS PDZ domain (Figure 6C).
Figure 6.
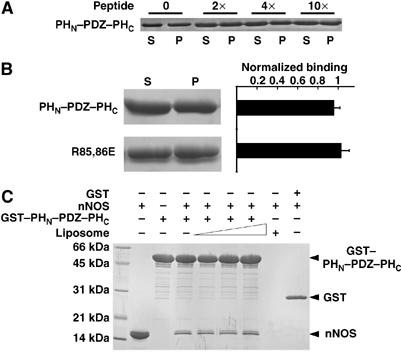
The PDZ ligands do not affect lipid binding of the PHN–PDZ–PHC tandem. (A) Addition of excess amounts (up to ∼10 equivalents) of a PDZ-binding peptide to the liposome-binding assay buffer did not alter the lipid-binding capacity of the PHN–PDZ–PHC tandem. The amino-acid sequence of the PDZ ligand peptide is: KLSSIESDV. (B) Mutation of the two positively charged residues (Arg85 and Arg86) in the carboxyl group-binding loop of the target recognition groove of the PDZ domain did not change lipid-binding capacity of the PHN–PDZ–PHC tandem. (C) Binding of lipids to the PHN–PDZ–PHC tandem does not change the binding of PDZ domain to its ligand, the nNOS PDZ domain. Three different liposome concentrations (0.4, 0.6, and 0.8 mg/ml) were tested in this assay.
Discussion
As one of the most abundant protein modules in the mammalian genome, PH domains have been extensively studied both structurally and functionally (Schultz et al, 1998b; Lemmon and Ferguson, 2000). PH domains are generally regarded as lipid-binding modules capable of targeting PH domain proteins to the cell membrane or detecting lipid signals generated in cellular signaling processes. A recent genome-wide analysis of all yeast PH domains suggested that only a minor population of PH domains binds to lipids specifically. Thus, the functions of the majority of PH domains are unknown (Yu et al, 2004). Protein domain searching using the SMART program (Schultz et al, 1998b) identifies several proteins, for example, syntrophins, PLCγ1, myosin X (Berg et al, 2000), and Rho kinases, that contain a split PH domain (i.e. the PH domain contains one or more other intact protein modules in the middle of its primary sequence). The significance of split PH domains has not been studied until a very recent report by van Rossum et al (2005), suggesting that the C-terminal half of the PLCγ1 PH domain may be complemented by a hidden partial PH domain-like segment in TRPC3. It was suggested that partial PH domains exists in many proteins (van Rossum et al, 2005). Detailed structural studies of split PH domains are pressingly needed to understand the novel functions of this unique subgroup of PH domain proteins.
As one of the major findings in this study, we found that the split PH domain of α-syntrophin adopts an intramolecular, canonical PH fold. We also showed that the PDZ domain insertion does not have significant impact on the structure of the split PH domain. We further showed that the two halves of the split PH domain, even in the absence of covalent linkage, can autonomously fold into a native-like PH domain structure. In contrast, the isolated partial fragments of the split PH domain of α-syntrophin are both unfolded, but soluble in solution. Our data indicate that partial PH domains may stably exist in proteins, and such partial PH domains easily escape detection by conventional computer algorithms. Interaction of the two complementary halves of a PH domain induces de novo folding of the domain. Thus, our study provides a structural basis for potential interactions of two hidden partial PH domains within one protein or between two different proteins. Our study also suggests that the intramolecular, two half-PH domain complementation dominates the assembly of the split PH domain of α-syntrophin. If this is also the case for the split PH domain of PLCγ1, the interaction between PLCγ1–PHC and the hypothetical partial PH domain of TRPC3 suggested by van Rossum et al (2005) requires a prior dissociation of PLCγ1–PHC from PLCγ1–PHN. If such intermolecular PH domain assembly indeed occurs, a key question is whether and how such a process is regulated.
Another key finding of this work is that the PHN–PDZ–PHC tandem functions as a supramodule with distinctly higher avidity in binding to lipids when compared to the isolated PHN–PHC and PDZ domains. The simplest explanation for the high avidity of the PHN–PDZ–PHC supramodule is the synergism afforded by the two weak lipid-binding domains (the split PH domain and the PDZ domain) covalently connected to each other. It is important to note that the binding between inositol phospholipids and the PHN–PDZ–PHC tandem requires the membrane bilayer and the hydrophobic tails of the lipids, as soluble head groups showed no detectable binding to he PHN–PDZ–PHC tandem. Presumably, anchoring of phosphoinositides onto the membrane bilayer is entropically favorable for the lipid molecules binding to the bidentate PHN–PDZ–PHC tandem. The interaction between PDZ domains and lipids is an emerging concept. The molecular basis of lipid/PDZ interaction is largely unknown, and this is an important area in the future research. It is interesting to note that the lipid-binding affinity of the PHN–PDZ–PHC tandem also depends on the position of the PDZ domain insertion. Furthermore, the canonical protein/peptide target-binding site of the PDZ domain does not overlap with the lipid-binding site of the domain. Many of the PDZ domain-binding proteins are membrane proteins (Sheng and Sala, 2001; Zhang and Wang, 2003). One can envision that the coordinated actions of lipid binding by the PHN–PDZ–PHC tandem and the PDZ domain-mediated interactions with membrane proteins can target syntrophins very efficiently to specialized membrane domains, such as neuromuscular junctions and astroglial endfeet. In a broader sense, the lipid- (and potentially protein-) binding properties of many PH domains could be influenced by other protein domains both intra- and intermolecularly. It is perhaps not surprising that many PH domains do not bind to lipids in their isolated forms.
Materials and methods
Protein expression and purification
The PHN–PDZ–PHC tandem (residues 2–264), the joint PHN–PHC domain (residues 2–79 and 165–264), and the PDZ domain (residues 80–164) of mouse α-syntrophin were cloned into a modified version of pET32a vector, respectively (Feng et al, 2002). The protease-cleavable form of the PHN–PHC domain was constructed by insertion of a protease 3C recognition sequence at the joint site of the two halves of the PH domain. Point mutations of the PHN–PDZ–PHC tandem (R16,30E, R244E, and R85,86S) were created by PCR-based mutagenesis. The PDZ–PHN–PHC and PHN–PHC–PDZ mutants were constructed, respectively, by connecting the PDZ domain and joint PHN–PHC domain with a 10-residue ‘Gly–Ser–Gly–Gly–Ser–Gly–Gly–Ser–Gly–Ser' linker. Bacterial cells harboring each fusion protein expression plasmid were grown at 37°C, and protein expression was induced by isopropyl-β-D-thiogalactoside at 16°C overnight. Uniformly 15N- and 15N/13C-labeled proteins were prepared by growing bacteria in M9 medium containing 15NH4Cl with or without 13C6-glucose. The His-tagged fusion proteins were purified under native conditions using a Ni-NTA agarose (Qiagen) affinity chromatography. The remaining small amount of contaminant proteins was removed by size-exclusion chromatography.
NMR structure determination
NMR samples contained ∼1.0 mM of the PHN–PDZ–PHC tandem in 100 mM potassium phosphate, pH 7.0, in 90% H2O/10% D2O or 99.9% D2O. NMR spectra were acquired at 30 °C on a Varian Inova 750 MHz spectrometer equipped with an actively z-gradient shielded triple resonance probe. Backbone and side-chain resonance assignments of the protein were obtained by standard heteronuclear correlation experiments (Bax and Grzesiek, 1993; Kay and Gardner, 1997). The stereo-specific assignments of the Val and Leu methyl groups were obtained using a 10% 13C-labeled sample (Neri et al, 1989). Approximate interproton distance restraints were derived from NOESY spectra (a 1H 2D homonuclear NOESY, a 15N-separated NOESY, and a 13C-separated NOESY). NOEs were grouped into three distance ranges: 1.8–2.7 Å (1.8–2.9 Å for NOEs involving NH protons), 1.8–3.3 Å (1.8–3.5 Å for NOEs involving NH protons), and 1.8–5.0 Å, corresponding to strong, medium, and weak NOEs, respectively. Hydrogen bonding restraints were generated from the standard secondary structure of the protein based on the NOE patterns and backbone secondary chemical shifts. Backbone dihedral angle restraints (φ and ψ angles) were derived from the chemical shift analysis program TALOS (Cornilescu et al, 1999). Structures were calculated using the program CNS (Brunger et al, 1998). Figures were generated using MOLMOL (Koradi et al, 1996), MOLSCRIPT (Kraulis, 1991), and Raster3D (Merritt and Murphy, 1994).
Lipid-binding assay
Brain lipid extracts (Folch fraction I, Sigma B1502) were resuspended at 2 mg/ml in a buffer containing 20 mM HEPES, pH 7.4, 150 mM NaCl, and 1 mM DTT. The protein sample (5 μM) was incubated with 0.6 mg/ml liposomes in 40 μl of buffer for 15 min at room temperature and then spun at 65 000 g for 15 min at 4°C in a Beckman TLA100.1 rotor. The supernatants were removed for determination of proteins not bound to liposomes. The pellets were washed twice with the same buffer and brought up to the same volume as the supernatant. The supernatant and the pellet proteins were subjected to SDS–PAGE and visualized by Coomassie blue staining.
Phosphatidylcholine, phosphatidylserine, and PI(3,5)P2 were purchased from Avanti Lipids (>99% pure). Large unilamellar vesicles (LUV) were generated by mixing di(dibromostearoyl) phosphatidylcholine and dipalmitoyl phosphatidyl-L-serine dissolved in chloroform in a 4:1 molar ratio. Addition of PI (3,5)P2 was counteracted by reduction of equal moles of PC. Mixed lipids were freeze dried, and resuspended in 50 mM Tris (pH 7.4) and 100 mM NaCl at 4.0 mM. Hydrated lipids were subjected to at least 10 cycles of freeze–thawing in liquid nitrogen, followed by 1 min bath sonication and extrusion through a 0.1 μm filter with a lipid extruder (Avanti). Purified PHN–PDZ–PHC was labeled with CPM (Molecular Probes) containing single Cys (Cys232) at the end of β6 of the split PH domain (Cys17 and Cys219 were substituted with Ser). PHN–PDZ–PHC was labeled with 10-fold molar excess of CPM in 50 mM Tris (pH 7.4) and 100 mM NaCl at 4°C. The reaction mixture was cleared by centrifugation, the excess CPM was removed by PD-10 column (Amersham Pharmacia). For constant-density PIP2 titrations, lipid vesicles were titrated into a 0.6 ml solution containing 0.3 μM CPM-labeled PHN–PDZ–PHC in 50 mM Tris (pH 7.4), 100 mM NaCl. The sample was excited at 386 nm, and emission was detected at 470 nm. Data from binding experiments were fit by nonlinear least-squares methods using Prism 4 from GraphPad.
Lipid strip assay
The PIP strips (Echelon, P-6001) were blocked by 3% fatty acid-free BSA in TBS-T buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1%Tween-20) at room temperature for 1 h. The strips were then incubated with 0.5 μg/ml of PHN–PDZ–PHC at 4°C overnight, washed five times in TBS-T buffer and subsequently incubated for 1 h with the pan-specific syntrophin antibody SYN1351 (1:8000) (Froehner et al, 1987) at room temperature. The signals were generated using the enhanced chemiluminescence detection kit (Perkin-Elmer).
Coordinates
The atomic coordinates have been deposited in the Protein Data Bank under access numbers of 1Z87, 2ADZ, and 1Z86 corresponding to the PHN–PDZ–PHC tandem, the joined PHN–PHC domain, and the isolated PDZ domain, respectively. The chemical shift assignments of the three proteins have been deposited in the BioMagResBank.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table 1
Acknowledgments
This work was supported by grants from the Research Grants Council of Hong Kong, the Human Frontier Science Program, and the Philip Morris Inc., USA, to MZ, and from the National Institutes of Health to SCF. The NMR spectrometer used in this work was purchased with funds donated to the Biotechnology Research Institute by the Hong Kong Jockey Club. MZ was a recipient of the Croucher Foundation Senior Research Fellow award.
References
- Adams ME, Butler MH, Dwyer TM, Peters MF, Murnane AA, Froehner SC (1993) Two forms of mouse syntrophin, a 58 kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron 11: 531–540 [DOI] [PubMed] [Google Scholar]
- Adams ME, Kramarcy N, Krall SP, Rossi SG, Rotundo RL, Sealock R, Froehner SC (2000) Absence of alpha-syntrophin leads to structurally aberrant neuromuscular synapses deficient in utrophin. J Cell Biol 150: 1385–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams ME, Mueller HA, Froehner SC (2001) In vivo requirement of the alpha-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. J Cell Biol 155: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn AH, Kunkel LM (1995) Syntrophin binds to an alternatively spliced exon of dystrophin. J Cell Biol 128: 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn AH, Yoshida M, Anderson MS, Feener CA, Selig S, Hagiwara Y, Ozawa E, Kunkel LM (1994) Cloning of human basic A1, a distinct 59-kDa dystrophin-associated protein encoded on chromosome 8q23–24. Proc Natl Acad Sci USA 91: 4446–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht DE, Froehner SC (2002) Syntrophins and dystrobrevins: defining the dystrophin scaffold at synapses. Neurosignals 11: 123–129 [DOI] [PubMed] [Google Scholar]
- Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26: 131–138 [Google Scholar]
- Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE (2000) Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci 113 (Part 19): 3439–3451 [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS (1996) Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell 84: 757–767 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buechler C, Boettcher A, Bared SM, Probst MC, Schmitz G (2002) The carboxyterminus of the ATP-binding cassette transporter A1 interacts with a beta2-syntrophin/utrophin complex. Biochem Biophys Res Commun 293: 759–765 [DOI] [PubMed] [Google Scholar]
- Chang J-S, Kim S-K, Kwon T-K, Bae SS, Min DS, Lee YH, Kim S-O, Seo J-K, Choi JH, Suh P-G (2005) Pleckstrin homology domains of phospholipase C-{gamma}1 directly interact with {beta}-tubulin for activation of phospholipase C-{gamma}1 and reciprocal modulation of {beta}-tubulin function in microtubule assembly. J Biol Chem 280: 6897–6905 [DOI] [PubMed] [Google Scholar]
- Chang J-S, Seok H, Kwon T-K, Min DS, Ahn B-H, Lee YH, Suh J-W, Kim J-W, Iwashita S, Omori A, Ichinose S, Numata O, Seo J-K, Oh Y-S, Suh P-G (2002) Interaction of elongation factor-1alpha and pleckstrin homology domain of phospholipase C-gamma 1 with activating its activity. J Biol Chem 277: 19697–19702 [DOI] [PubMed] [Google Scholar]
- Chockalingam PS, Gee SH, Jarrett HW (1999) Pleckstrin homology domain 1 of mouse alpha 1-syntrophin binds phosphatidylinositol 4,5-bisphosphate. Biochemistry 38: 5596–5602 [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Bushweller JH (2004) Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. J Am Chem Soc 126: 16259–16266 [DOI] [PubMed] [Google Scholar]
- Connors NC, Adams ME, Froehner SC, Kofuji P (2004) The potassium channel Kir4.1 associates with the dystrophin–glycoprotein complex via alpha-syntrophin in glia. J Biol Chem 279: 28387–28392 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Feng W, Fan JS, Jiang M, Shi YW, Zhang M (2002) PDZ7 of glutamate receptor interacting protein binds to its target via a novel hydrophobic surface area. J Biol Chem 277: 41140–41146 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1994) Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell 79: 199–209 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA (1997) High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem 272: 8474–8481 [DOI] [PubMed] [Google Scholar]
- Froehner SC, Murnane AA, Tobler M, Peng HB, Sealock R (1987) A postsynaptic Mr 58,000 (58 K) protein concentrated at acetylcholine receptor-rich sites in Torpedo electroplaques and skeletal muscle. J Cell Biol 104: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Kojima T, Kabayama H, Mikoshiba K (1996) Mutation of the pleckstrin homology domain of Bruton's tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J Biol Chem 271: 30303–30306 [DOI] [PubMed] [Google Scholar]
- Fushman D, Cahill S, Lemmon MA, Schlessinger J, Cowburn D (1995) Solution structure of pleckstrin homology domain of dynamin by heteronuclear NMR spectroscopy. Proc Natl Acad Sci USA 92: 816–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee SH, Madhavan R, Levinson SR, Caldwell JH, Sealock R, Froehner SC (1998) Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. J Neurosci 18: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Cuenda A, Spillantini MG, Thomas GM, Buee-Scherrer V, Cohen P, Goedert M (1999) Stress-activated protein kinase-3 interacts with the PDZ domain of alpha1-syntrophin. A mechanism for specific substrate recognition. J Biol Chem 274: 12626–12631 [DOI] [PubMed] [Google Scholar]
- Hosaka Y, Yokota T, Miyagoe-Suzuki Y, Yuasa K, Imamura M, Matsuda R, Ikemoto T, Kameya S, Takeda S (2002) Alpha1-syntrophin-deficient skeletal muscle exhibits hypertrophy and aberrant formation of neuromuscular junctions during regeneration. J Cell Biol 158: 1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C (2004) Novel techniques for weak alignment of proteins in solution using chemical tags coordinating lanthanide ions. J Biomol NMR 29: 339–349 [DOI] [PubMed] [Google Scholar]
- Kachinsky AM, Froehner SC, Milgram SL (1999) A PDZ-containing scaffold related to the dystrophin complex at the basolateral membrane of epithelial cells. J Cell Biol 145: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameya S, Miyagoe Y, Nonaka I, Ikemoto T, Endo M, Hanaoka K, Nabeshima Y, Takeda S (1999) Alpha1-syntrophin gene disruption results in the absence of neuronal-type nitric-oxide synthase at the sarcolemma but does not induce muscle degeneration. J Biol Chem 274: 2193–2200 [DOI] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA (1998) Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem 273: 30497–30508 [DOI] [PubMed] [Google Scholar]
- Kay LE, Gardner KH (1997) Solution NMR spectroscopy beyond 25 kDa. Curr Opin Struct Biol 7: 722–731 [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55 [DOI] [PubMed] [Google Scholar]
- Kramarcy NR, Vidal A, Froehner SC, Sealock R (1994) Association of utrophin and multiple dystrophin short forms with the mammalian M(r) 58,000 dystrophin-associated protein (syntrophin). J Biol Chem 269: 2870–2876 [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM (2000) Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J 350 (Part 1): 1–18 [PMC free article] [PubMed] [Google Scholar]
- Leonoudakis D, Conti LR, Anderson S, Radeke CM, McGuire LM, Adams ME, Froehner SC, Yates JR III, Vandenberg CA (2004) Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)-associated proteins. J Biol Chem 279: 22331–22346 [DOI] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG (2000) Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell 6: 385–394 [DOI] [PubMed] [Google Scholar]
- Lumeng C, Phelps S, Crawford GE, Walden PD, Barald K, Chamberlain JS (1999) Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat Neurosci 2: 611–617 [DOI] [PubMed] [Google Scholar]
- Macias MJ, Musacchio A, Ponstingl H, Nilges M, Saraste M, Oschkinat H (1994) Structure of the pleckstrin homology domain from beta-spectrin. Nature 369: 675–677 [DOI] [PubMed] [Google Scholar]
- Merritt E, Murphy M (1994) Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr D50: 869–873 [DOI] [PubMed] [Google Scholar]
- Mortier E, Wuytens G, Leenaerts I, Hannes F, Heung MY, Degeest G, David G, Zimmermann P (2005) Nuclear speckles and nucleoli targeting by PIP(2)-PDZ domain interactions. EMBO J 24: 2556–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely JD, Amiry-Moghaddam M, Ottersen OP, Froehner SC, Agre P, Adams ME (2001) Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proc Natl Acad Sci USA 98: 14108–14113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D, Szyperski T, Otting G, Senn H, Wuthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28: 7510–7516 [DOI] [PubMed] [Google Scholar]
- Ou Y, Strege P, Miller SM, Makielski J, Ackerman M, Gibbons SJ, Farrugia G (2003) Syntrophin gamma 2 regulates SCN5A gating by a PDZ domain-mediated interaction. J Biol Chem 278: 1915–1923 [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA (2005) A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell 17: 181–191 [DOI] [PubMed] [Google Scholar]
- Piluso G, Mirabella M, Ricci E, Belsito A, Abbondanza C, Servidei S, Puca AA, Tonali P, Puca GA, Nigro V (2000) Gamma1- and gamma2-syntrophins, two novel dystrophin-binding proteins localized in neuronal cells. J Biol Chem 275: 15851–15860 [DOI] [PubMed] [Google Scholar]
- Pintacuda G, Moshref A, Leonchiks A, Sharipo A, Otting G (2004) Site-specific labelling with a metal chelator for protein-structure refinement. J Biomol NMR 29: 351–361 [DOI] [PubMed] [Google Scholar]
- Rameh LE, Arvidsson A-K, Carraway KL III, Couvillon AD, Rathbun G, Crompton A, VanRenterghem B, Czech MP, Ravichandran KS, Burakoff SJ, Wang D-S, Chen C-S, Cantley LC (1997) A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem 272: 22059–22066 [DOI] [PubMed] [Google Scholar]
- Schultz J, Hoffmuller U, Krause G, Ashurst J, Macias MJ, Schmieder P, Schneider-Mergener J, Oschkinat H (1998a) Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nat Struct Biol 5: 19–24 [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998b) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C (2001) Pdz domains and the organization of supramolecular complexes. Annu Rev Neurosci 24: 1–29 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yoshida M, Ozawa E (1995) Mammalian alpha 1- and beta 1-syntrophin bind to the alternative splice-prone region of the dystrophin COOH terminus. J Cell Biol 128: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CC, Deak M, Alessi DR, van Aalten DM (2002) High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr Biol 12: 1256–1262 [DOI] [PubMed] [Google Scholar]
- Touhara K, Inglese J, Pitcher J, Shaw G, Lefkowitz R (1994) Binding of G protein beta gamma-subunits to pleckstrin homology domains. J Biol Chem 269: 10217–10220 [PubMed] [Google Scholar]
- van Rossum DB, Patterson RL, Sharma S, Barrow RK, Kornberg M, Gill DL, Snyder SH (2005) Phospholipase Cgamma1 controls surface expression of TRPC3 through an intermolecular PH domain. Nature 434: 99–104 [DOI] [PubMed] [Google Scholar]
- Yang B, Ibraghimov-Beskrovnaya O, Moomaw CR, Slaughter CA, Campbell KP (1994) Heterogeneity of the 59-kDa dystrophin-associated protein revealed by cDNA cloning and expression. J Biol Chem 269: 6040–6044 [PubMed] [Google Scholar]
- Yoon HS, Hajduk PJ, Petros AM, Olejniczak ET, Meadows RP, Fesik SW (1994) Solution structure of a pleckstrin-homology domain. Nature 369: 672–675 [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13: 677–688 [DOI] [PubMed] [Google Scholar]
- Zhang M, Wang W (2003) Organization of signaling complexes by PDZ-domain scaffold proteins. Accounts Chem Res 36: 530–538 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J (2002) PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell 9: 1215–1225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table 1


