Figure 1.
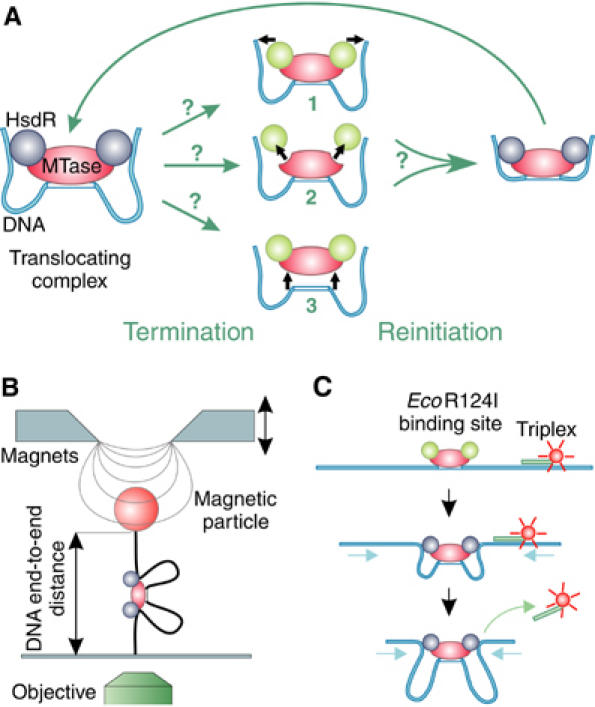
Schematic representation of termination and reinitiation of translocation by Type I RE and sketches of the two methods to measure translocation. (A) Three possible mechanisms for the termination of translocation by a loop-forming enzyme: (1) release of the translocated DNA by the HsdR subunits, with the enzyme complex remaining bound on the DNA; (2) disassembly of the enzyme by dissociation of the HsdR subunits from the MTase core unit; and (3) complete dissociation of the enzyme complex from the DNA. Following termination of translocation, reinitiation via the formation of an initial loop can occur; the mechanism utilized will depend on the termination route. Static HsdR molecules are drawn in green and translocating molecules in blue. (B) Magnetic tweezers. A DNA molecule with a single EcoR124I binding site is anchored between a magnetic bead and a glass slide. A pair of magnets is used to stretch the DNA molecule with different forces. Translocation of EcoR124I can be seen as a decrease in the DNA end-to-end distance, which is measured using video microscopy. (C) Triplex displacement assay. A tetramethylrhodamine-labeled triplex is attached at specific distances from the EcoR124I binding site. Translocation leads to displacement of the triplex resulting in an increase of fluorescence, which is monitored with a stopped-flow apparatus.
