Figure 1.
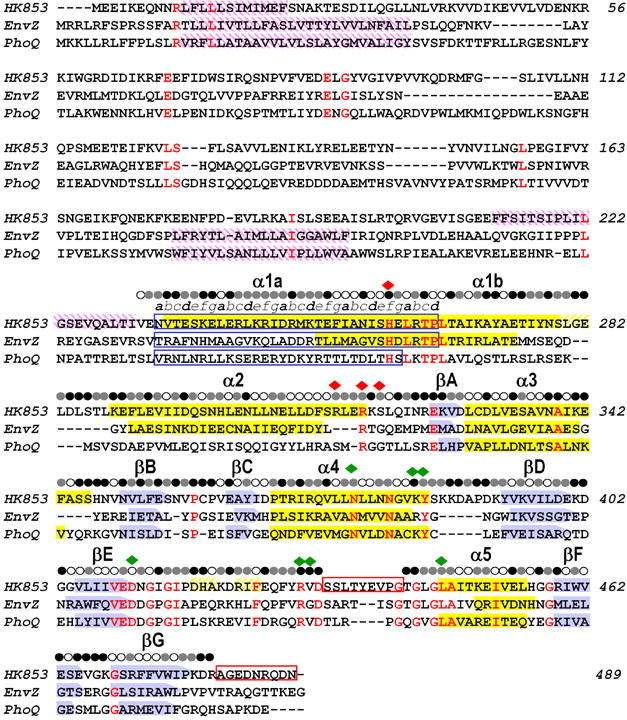
Sequence alignment of TM0853, EnvZ and PhoQ HKs. The three amino-acid sequences are aligned based on their structures. β sheets are shown as blue arrows and α helices as yellow filled boxes. Transmembrane regions predicted by the DAS program (Cserzo et al, 1997) are shown as purple intermediate shading and coiled-coil motifs predicted by LEARNCOIL program (Singh et al, 1998) are enclosed in blue boxes showing the helical position from a to g. Disordered regions are enclosed in red boxes. Residues identical in all three sequences are colored in red. The solvent accessibility of the HK853–CD is indicated for each residue by an open circle if the fraction solvent accessibility is >0.4, a half-filled circle if it is 0.1–0.4 and a filled circle if it is <0.1. Residues that interact with the ADPβN and the sulfate ion in HK853–CD are indicated by green and red diamonds, respectively.
