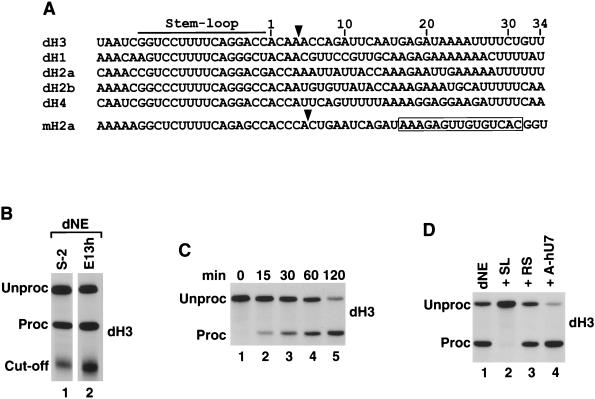
FIG. 1.
dH3 pre-mRNA is efficiently processed in nuclear extracts from Drosophila cells. (A) Sequence alignment of the 3′ end region of all five Drosophila histone pre-mRNAs and the mouse H2a-614 pre-mRNA. The stem-loop structure is indicated with the line, and the U7 binding site in the mouse H2a pre-mRNA is boxed. The known cleavage site used in mammalian processing and the deduced cleavage site used in Drosophila processing (see below) are indicated by the arrow. The nucleotides are numbered beginning from the first nucleotide after the stem-loop. (B) In vitro processing of radiolabeled dH3 pre-mRNA in dNE from S-2 cells (lane 1) and 0- to 13-h-old embryos (lane 2). The reaction was carried out for 2 h at 22°C, and RNA was resolved by electrophoresis in a 6% denaturing gel. “Unproc” and “Proc” correspond to the input pre-mRNA and the upstream cleavage product (mRNA), respectively. The band indicated as the “Cut-off” corresponds to the 3′ cleavage product. (C) Time course of Drosophila 3′ end processing. The dH3 pre-mRNA was incubated for the indicated times in nuclear extract from S-2 cells. (D) The dH3 pre-mRNA was incubated in the S-2 nuclear extract in the presence of excess of 26-nucleotide stem-loop RNA (SL, lane 2), reverse stem RNA (RS, lane 3), or a 2′ O-methyl oligonucleotide complementary to the 5′ end of human U7 snRNA (A-hU7, lane 4). The control processing reaction is shown in lane 1.