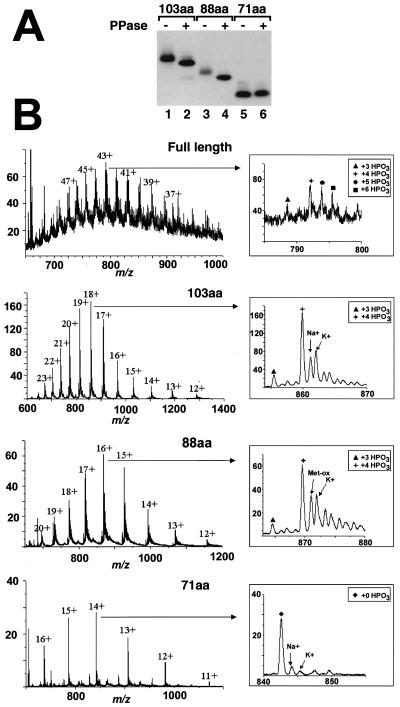
FIG. 8.
dSLBP contains four phosphates on the C-terminal region. (A) The deletion mutants of dSLBP shown in Fig. 5B were expressed by using the baculovirus system and purified. Aliquots of each variant were treated with PPase, and the untreated (lanes 1, 3, and 5) and dephosphorylated (lanes 2, 4, and 6) proteins were separated by gel electrophoresis and then detected by Western blotting. The mobility of the 103 (lanes 1 and 2)- and 88 (lanes 3 and 4)-amino-acid dSLBPs was increased after PPase treatment, whereas the mobility of the 71-amino-acid protein containing only the RBD was unchanged (lanes 5 and 6). (B) The full-length dSLBP and the 103-, 88-, and 71-amino-acid dSLBPs expressed in insects cells were analyzed by mass spectrometry as described in Materials and Methods. The charge/mass (m/z) spectrum is shown, and the charges on the individual clusters of peaks are indicated. The insets show a blowup of the most abundant ion. The various phosphorylated forms and proteins associated with sodium or potassium ions are indicated. The major peaks in the full-length, 103- and 88-amino-acid dSLBPs correspond to a molecular mass of 320 Da higher than that predicted from the amino acid sequence, corresponding to the four phosphate groups. The two satellite peaks indicated by the circle and square in the full-length dSLBP correspond to additional partially phosphorylated molecules containing five and six phosphates, respectively, whereas the peak indicated by the triangle corresponds to a protein containing three phosphates. The major peak in the 71-amino-acid SLBP corresponds to the molecular weight predicted from the amino acid sequence, indicating that the RBD is not phosphorylated.