Abstract
Cell specification in the nervous system requires patterning genes dictating spatio-temporal coordinates as well as fate determinants. In the case of neurons, which are controlled by the family of proneural transcription factors, binding specificity and patterned expression trigger both differentiation and specification. In contrast, a single gene, glide cell deficient/glial cell missing (glide/gcm), is sufficient for all fly lateral glial differentiation. How can different types of cells develop in the presence of a single fate determinant, that is, how do differentiation and specification pathways integrate and produce distinct glial populations is not known. By following an identified lineage, we here show that glia specification is triggered by high glide/gcm expression levels, mediated by cell-specific protein-protein interactions. Huckebein (Hkb), a lineage-specific factor, provides a molecular link between glide/gcm and positional cues. Importantly, Hkb does not activate transcription; rather, it physically interacts with Glide/Gcm thereby triggering its autoregulation. These data emphasize the importance of fate determinant cell-specific quantitative regulation in the establishment of cell diversity.
Keywords: autoregulation, Drosophila melanogaster, glia specification, Glide/Gcm, Huckebein
Introduction
Cell specification requires the activity of patterning genes and cell fate determinants. Proneural genes trigger both neuronal differentiation and specification, and lack of any of them leads to the absence of identified neuronal subpopulations (Skeath, 1999; Bertrand et al, 2002). Multiple features allow proneural transcription factors of the basic helix–loop–helix (bHLH) family to generate diversity. Their expression is controlled by patterning genes along dorso/ventral (D/V), antero/posterior (A/P) and temporal axes; moreover, each factor is able to bind specific E boxes (Powell et al, 2004) and cofactors (Ramain et al, 2000; Lee and Pfaff, 2003; zur Lage et al, 2004).
A functional nervous system also relies on different types of glial cells (Ito et al, 1995; Bossing et al, 1996b; Schmidt et al, 1997; Van De Bor and Giangrande, 2002; Rowitch, 2004). Patterning genes have been shown to define broad gliogenic territories (for reviews see Skeath and Thor, 2003; Rowitch, 2004; Soustelle and Giangrande, 2005); however, the molecular cues underlying glia specification at the cellular level within and among these territories are still poorly understood. Knowing the gene that induces glial differentiation and being able to follow identified lineages make it possible to tackle this issue in Drosophila melanogaster. glide/gcm gene (referred throughout the text as gcm, for the sake of simplicity) is expressed and required in all lateral glial cells of the fly central nervous system (CNS) (Hosoya et al, 1995; Jones et al, 1995; Vincent et al, 1996). The glia to neuron transformation observed in gcm embryos and the fact that gcm ectopic expression leads to the differentiation of additional glial cells at the expense of endogenous developmental programs identify gcm as a molecular switch during cell fate decision (Hosoya et al, 1995; Jones et al, 1995; Vincent et al, 1996; Akiyama-Oda et al, 1998; Bernardoni et al, 1998).
These findings raise the question of how are different types of glial cells specified. In particular, does glia specification depend on the single gcm fate determinant or on a parallel pathway? Also, how and where do cell differentiation and specification integrate in order to produce the precise array of glial cells that characterize the nervous system? By analyzing the neuroglioblast 1-1 abdominal (NGB1-1A) lineage of the fly CNS, we here show that integration occurs at the level of the Gcm protein, the amount of which is controlled by Huckebein (Hkb). We also show for the first time that Hkb, a repressor controlling terminal patterning in the fly embryo (Weigel et al, 1990; Bronner and Jackle, 1991; Bronner et al, 1994), does not work as a transcription factor. Rather, its direct interaction with Gcm triggers gcm autoregulation and thereby promotes differentiation of a specific type of subperineural glia (SPG). Thus, we demonstrate the importance of fate determinant quantitative regulation in cell specification. hkb represents one of the lineage-specific factors controlled by patterning genes in the nervous system (Skeath, 1999). By identifying its mode of action, we provide the molecular link between patterning and specification here.
Results
Hkb is necessary for gcm expression in the NGB1-1A lineage
Each segment of the fly embryonic ventral cord contains about 60 stereotypically organized lateral glial cells (Figures 1A and 2A), most of which arise from neuroglioblasts (NGBs), mixed precursors producing both neurons and glia (Bossing et al, 1996b; Schmidt et al, 1997). In the absence of Gcm, glia are absent (Figure 2B) and transform into neurons (Hosoya et al, 1995; Jones et al, 1995; Akiyama et al, 1996; Vincent et al, 1996; Schreiber et al, 1997; Bernardoni et al, 1998; Miller et al, 1998; Akiyama-Oda et al, 1999). This phenotype is also observed in hkb embryos (Figure 1C), but restricted to lateral glia derived from NGB1-1A (Bossing et al, 1996a). These data suggest that Hkb and Gcm work in the same pathway in NGB1-1A, prompting us to use this lineage as a model to understand the bases of glia specification.
Figure 1.
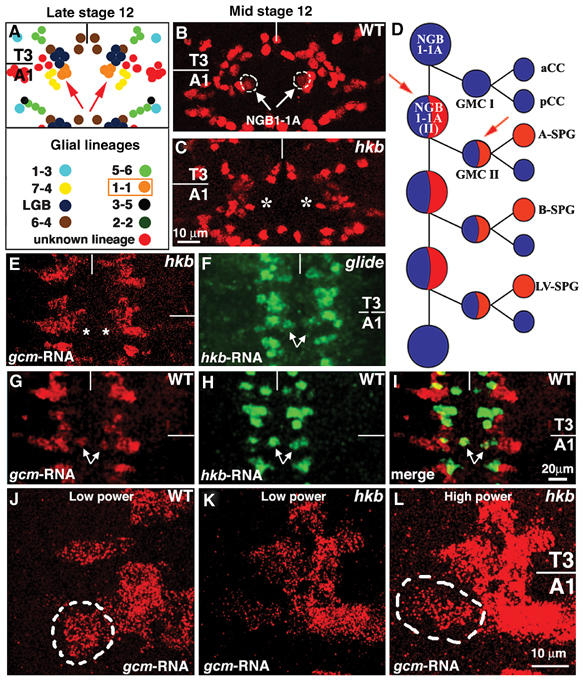
hkb controls glial differentiation in the NGB1-1A lineage. Unless otherwise specified, panels in this and in following figures show ventral views of the embryonic ventral cord; T3 and A1 indicate, respectively, third thoracic and first abdominal segments; anterior is to the top and the vertical line indicates the midline. (A) Schematic drawing of Repo labeled cells in T3 and A1 segments of a wild-type (WT)_ embryo at late stage 12. Glial subsets are identified by the expression of lineage-specific markers indicated by different colors. Symbols as in Ragone et al (2003). (B, C) Mid stage 12 embryos, WT (B) or hkb (C), labeled with glial-specific antibody anti-Repo. Arrows and dashed lines indicate NGB1-1A-derived glia. Asterisks in (C) indicate the absence of Repo labeling at the position normally taken by NGB1-1A-derived glia. (D) NGB1-1A lineage as proposed by Udolph et al (2001). Glial and neuronal potentials/components are indicated by red and blue, respectively. The first division of NGB1-1A gives rise to a ganglion mother cell (GMC I) that produces neurons (aCC and pCC), whereas the second division (NGB1-1A (II)) gives rise to a GMC (GMC II) that produces one neuron (n) and one glial cell (A-SPG). B-SPG and LV-SPG arise from later divisions. Red arrows indicate the two cells in which gcm mRNA is detected at stage 11. (E–L) Late stage 11 embryos. (E) hkb embryo labeled with gcm riboprobe (asterisks indicate the position normally taken by gcm expressing cells in the NGB1-1A lineage). (F) gcm embryo labeled with hkb riboprobe. (G–I) WT embryo labeled with gcm (G) and hkb (H) riboprobes. (I) Merge of (G and H). Arrows in (F–I) indicate cells of the NGB1-1A lineage. (J–L) WT (J) or hkb (K, L) embryos labeled with gcm riboprobe. Note the very low levels of gcm expression in mutant (K) compared to WT (J, dashed line) NGB1-1A lineage. (L) Same embryo as in (K) analyzed at high photomultiplicator power to amplify signal. Scale bars: 10 μm in (B, C, J–L) and 20 μm in (E–I).
Figure 2.
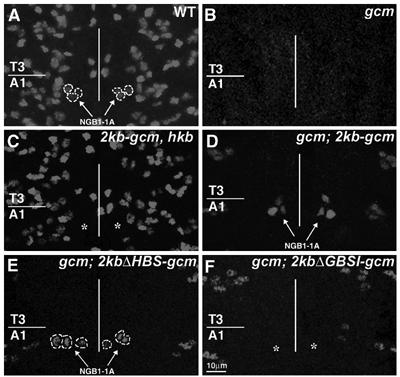
NGB1-1A-derived glial cells depend on 2kb-gcm promoter. Late stage 12 embryos labeled with anti-Repo. (A) NGB1-1A glia are shown in a wild type (WT) by arrows and dashed lines. (B) No Repo positive nuclei are present in a gcm embryo (gcm). (C) NGB1-1A glial cells are absent in hkb embryo carrying the 2kb-gcm transgene (see asterisks), but present in gcm embryo carrying the same (D) or 2kbΔHBS-gcm (E) transgenes (see arrows) (100% of the animals, n=15). (F) NGB1-1A glia are absent in gcm embryos carrying the 2kbΔGBSI-gcm transgene (40% of the animals, n=50), see asterisks. Note that several transgenes were analyzed for each construct. To take into account position effects, transgenes of comparable strength were used. Scale bar: 10 μm.
NGB1-1A lineage contains six to eight neurons and three glial cells of the SPG class (Broadus et al, 1995; Ito et al, 1995; Bossing et al, 1996b) (Figure 1A). NGB1-1A first produces aCC and pCC motoneurons and subsequently gives rise to several ganglion mother cells (GMCs), each producing a neuron and a glial cell that do not divide further (Udolph et al, 2001) (Figure 1D). hkb transcripts are first detected at the time gliogenesis starts (Chu-LaGraff et al, 1995) (Figures 1H and 3D–F) and colocalize with gcm RNA within the NGB1-1A lineage (Figures 1G–I and 3A–C, J–M). Expression of both mRNAs starts at stage 11 in a single, dividing, cell (Figures 3A–M). By late stage 11, two cells express gcm; one of them is somewhat larger, more apically located than the other, and corresponds to NGB1-1A (II) (Figure 3N). This cell expresses both gcm and hkb, whereas the basal, small, cell corresponds to ganglion mother cell II (GMC II) (see Figure 1D) and only expresses gcm (Figure 3N). Repo glial-specific marker (Campbell et al, 1994; Xiong et al, 1994; Halter et al, 1995) starts being detected in one cell of the lineage by mid stage 12 (Figure 1B), three cells per hemisegment being labeled at late stage 12 (Figures 1A and 2A).
Figure 3.
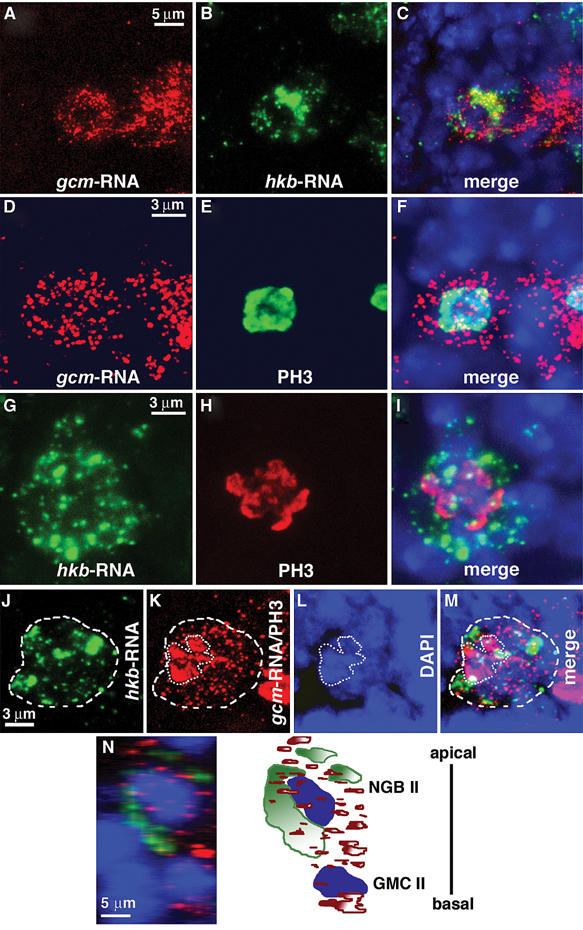
hkb and gcm are coexpressed in NGB1-1A (II). NG1-1A lineage multiple labelling in wild-type (WT) embryos. (A–C) Triple labeling: (A) gcm riboprobe, (B) hkb riboprobe, (C) is a merge showing DAPI (blue) nuclear labeling. (D–F) Triple labeling: (D) gcm riboprobe, (E) anti-PH3 mitotic marker, (F) is a merge showing DAPI. (G–I) Triple labeling: (G) hkb riboprobe, (H) anti-PH3, (I) is a merge showing DAPI. (J–M) quadruple labeling: (J) hkb riboprobe (dashed line), (K) anti-PH3/gcm riboprobe, (L) DAPI labeling (dotted line), (M) is a merge. In (K), the same secondary antibody was used to reveal gcm RNA (dashed line) and PH3 (dotted line) because primary antibodies used were raised in the same species, but subcellular localization of gcm RNA and PH3 enables to distinguish between the two stainings. (N) 90° rotation to show labeling along the Z-axis. Triple labeling at two cell stage: hkb riboprobe (green), gcm riboprobe (red) and DAPI (blue). hkb-RNA is localized only in NGB1-1A II, whereas gcm-RNA is localized both in NGB and in GMC II, as schematically represented in the right panel. Scale bars: 5 μm in (A–C and N) and 3 μm in (D–M).
As NGB1-1A glia are missing in hkb embryos (Figure 1C), we determined whether hkb controls gcm. Very low levels of gcm transcripts are indeed present in the NGB1-1A lineage of hkb embryos (Figures 1J–L), gcm expression remaining unaffected in the rest of the ventral cord. In contrast, NGB1-1A hkb expression is not affected by the absence of gcm (Figure 1F).
Previous analyses have allowed us to identify the first 2kb upstream of gcm as sufficient to drive gcm expression in NGB1-1A lineage (Ragone et al, 2003). When introduced into a gcm background, a transgene carrying the gcm coding sequences together with the 2kb upstream region (2kb-gcm) rescues differentiation of NGB1-1A derived glial cells (Figure 2D). In contrast, the same transgene does not rescue NGB1-1A glia when introduced into hkb embryos (Figure 2C). Thus, the 2kb promoter that drives NGB1-1A glia development requires Hkb.
Hkb does not directly activate gcm transcription
Hkb is known to bind specific sequences (Kuhnlein et al, 1997) and was first identified as a putative transcription factor based on the presence of a glutamine-rich domain (Bronner et al, 1994). More recently, it was shown that Hkb interacts with the Groucho corepressor and negatively regulates the expression of several target genes, such as snail and brachyenteron (Goldstein et al, 1999). Interestingly, we have found that Hkb-predicted open reading frame (ORF) does not contain the glutamine-rich region (putative activation domain), as further confirmed by the genome annotation (release 4.1, 2005) of the Genome Sequence Project. The published sequence contains a two-nucleotide deletion that changes the ORF in the N-terminal (Nt) region, whereas a third deletion of one nucleotide upstream of the Zn-finger motifs restores the correct ORF (Figure 4A). Thus, Hkb protein does contain three Zn-finger motifs in the C-terminal (Ct) region as well as the Groucho binding motif (FRPW) (Goldstein et al, 1999) (Figure 4B), but does not contain the putative activation domain.
Figure 4.
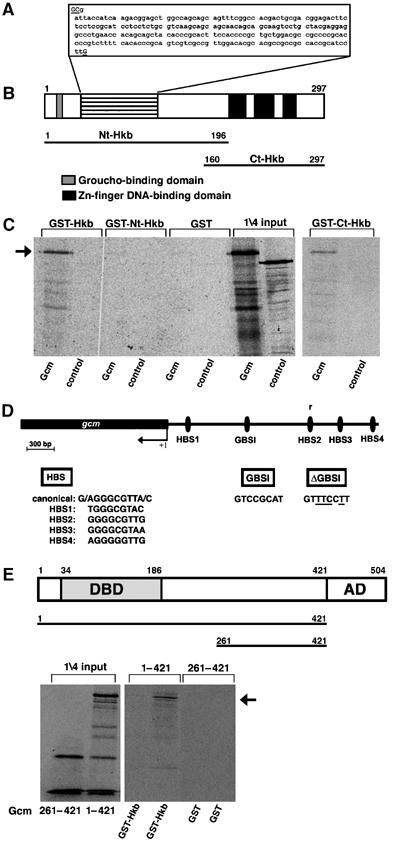
Hkb predicted ORF and interaction with Gcm. (A) Nucleotide sequence in the region indicated by horizontal lines in (B). Underlined nucleotides are absent in the published sequence (Bronner et al, 1994). (B) Organization of the predicted Hkb ORF. Boxes indicate the Zn-finger motifs (black) and the Groucho binding domain (gray) (Goldstein et al, 1999). Lines in (B) below the ORF indicate the Hkb truncated forms used to map interaction domains in GST pull-down assays. (C) Autoradiography of a pull-down assay using full length Gcm (amino acids 1–504) and GST–Hkb derivatives immobilized on glutathione-agarose beads. Binding of full-length Hkb protein (GST–Hkb) and Hkb C-terminal part (GST–Ct-Hkb) is indicated by an arrow. Luciferase (Luc) in vitro translated protein is used as a control. 1/4 of the input is shown in the right part of the panel. (D) Four Hkb binding sites (HBS1–4) are present in the 2kb-gcm promoter, one of which is in the opposite orientation (r). Canonical HBS represents the site identified by Kuhnlein et al (1997). GBSI indicates the Gcm binding site present in the 2kb-gcm promoter (Ragone et al, 2003). +1 indicates the transcription start site. (E) Gcm ORF: DBD indicates DNA binding domain, and AD, activation domain (see for a review Van De Bor and Giangrande, 2002). Lines below ORF indicate the in vitro translated products used to map interaction domains in GST pull-down assay shown in the bottom panel. Arrow indicates binding of GST–Hkb with translated Gcm 1-421 product. Note that GST pull-down assays entail a DNAse treatment, which eliminates possible DNA contamination.
Both genetic data and expression profile analysis prompted us to determine whether Hkb induces NGB1-1A glial differentiation by regulating gcm expression directly. Four Hkb putative binding sites (HBS) were identified in the 2kb-gcm promoter (Figure 4D), the region that requires Hkb. These sites bind purified recombinant GST–Hkb fusion protein (Figure 5A). Binding specificity was confirmed by using mutagenized primers, as a two-nucleotide mutation abolishes interaction with Hkb (Figure 5B). Furthermore, we challenged the Hkb–DNA complex with increasing amounts of specific and nonspecific DNA competitors. Only specific cold competitor displaces the GST–Hkb protein from labeled oligonucleotides (Figure 5C). When tested in CAT assays, however, none of the HBSs induces gene expression upon cotransfection with an Hkb expression vector (Figure 5D).
Figure 5.
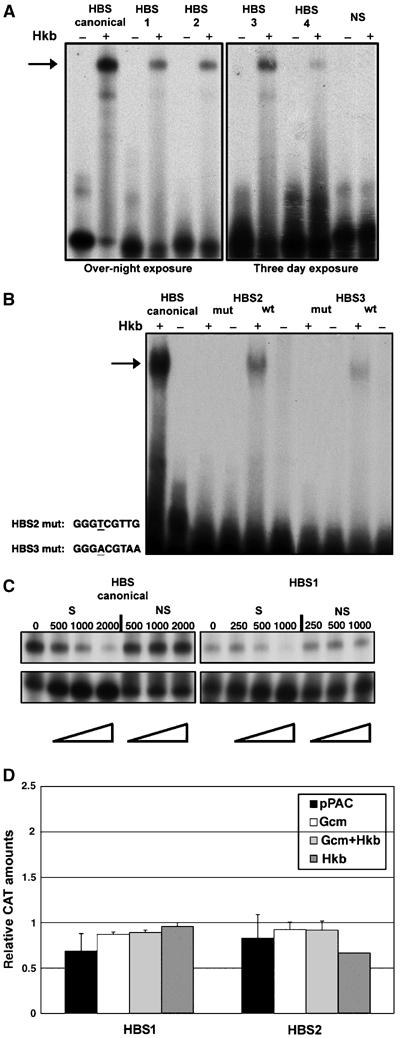
Hkb binds to its target sequences but does not act as a transcription factor. (A) Gel-shift assay showing DNA binding of a purified GST-–Hkb fusion protein (Hkb). Labeled 27-mers corresponding to each of the four HBSs, to the canonical HBS (Kuhnlein et al, 1997) or to nonspecific DNA (NS: GCATGGACCAACATTGACACCGCTTTG) were used in the assay. Binding is indicated by arrows. (B) Hkb binding is abolished when HBS2 or HBS3 carrying point mutations are used (HBS2mut and HBS3mut, respectively, mutant nucleotides underlined). Binding to the canonical site is shown as a positive control. Arrow indicates the position of bound 27mers. (C) Competition gel-shift assay on canonical HBS and HBS1. S and NS indicate specific and nonspecific cold competitors, respectively (X indicates folds of excess, 0 indicates the absence of competitor). (D) pBLCAT5 reporter constructs containing either HBS1 or HBS2 (HBS1 and HBS2) were cotransfected with one (pPAC, pPAC-Gcm or pPAC-Hkb) or two (pPAC-Gcm and pPAC-Hkb) expression vectors. CAT assay data were normalized by using control reporter vector pBLCAT5. Each bar represents the average of at least three measurements, and error bars indicate standard error.
Finally, we mutagenized the four HBSs present in the 2kb-gcm transgene (2kbΔHBS-gcm) and determined the ability of this mutant transgene to rescue glial cells of NGB1-1A lineage in gcm embryos. In line with the cotransfection data, the 2kbΔHBS-gcm transgene rescues NGB1-1A glia (Figure 2E), indicating that Hkb does not work as a transcription factor.
Hkb–Gcm interaction controls gcm positive autoregulation
It has been shown that Gcm positively and directly auto regulates in vitro, and that in vivo, this positive feedback loop requires cell-specific cofactors (Miller et al, 1998; Ragone et al, 2003). We speculated that Hkb represents one such factor and regulates gcm expression indirectly by binding the Gcm protein. Indeed, the 2kb-gcm promoter that is sufficient to drive rescue of NGB1-1A glia contains a Gcm binding site (GBS) (Miller et al, 1998; Ragone et al, 2003) (Figure 4D). We performed GST pull-down assays by using an Hkb–GST fusion expressed in bacteria and found that in vitro-translated Gcm binds GST–Hkb, but not GST alone (negative control) (Figure 4C). Binding specificity was further confirmed by using in vitro-translated Luciferase instead of Gcm (Figure 4C). Deletion analyses indicate that binding is mediated by Gcm Nt and HKB C-terminal (Ct-Hkb) regions (Figures 4C and E). Gcm–Hkb interactions were also confirmed in two-hybrid assay (Supplementary Figure 1).
We then determined the functional relevance of Hkb-Gcm interaction in DNA binding and CAT assays. The binding profile of Gcm Nt region (amino acids 1–261) to its site is modified by adding Hkb, which, on its own, does not bind GBSI (the site present in the 2kb-gcm promoter) (Figure 6A). An anti-Hkb, but not a nonspecific antibody, eliminates the band shift observed in the presence of Gcm and Hkb (Figure 6A), further demonstrating that Gcm–Hkb complex binds to GBS. Moreover, cotransfection assays using a reporter vector-containing GBSI show that Hkb does not trigger any detectable CAT activity (Figure 6B) and that combined Gcm and Hkb expression leads to two-fold increase of CAT activity, compared to that found with Gcm alone (Figure 6B). This effect is abolished upon GBSI mutagenesis (Figure 6B). Similar synergistic effects were obtained when a different GBS (site 1, present upstream of the 2kb promoter fragment; Miller et al, 1998; Ragone et al, 2003) was used in cotransfection assays as seen in Figure 6C. Nt-Hkb does not have any activation effect, whereas Ct-Hkb has almost the same effect as that observed with Hkb full-length protein, when cotransfected with Gcm (Figure 6B), in agreement with the pull-down data. Thus, the Zn-finger-containing region of Hkb triggers gcm autoregulation, even though this is not mediated by DNA binding, as Hkb does not bind GBS (Figure 6A). In addition, 2kbΔGBSI-gcm transgene that carries a mutated GBSI does not rescue NGB1-1A glia in a gcm background (Figure 2F). Finally, combined Gcm and Hkb expression does not trigger any detectable CAT activity when the reporter vector contains HBS (HBS1 and HBS2), reinforcing the idea that Gcm and Hkb do not act via Hkb-mediated transcription (Figure 5D). Our in vitro and in vivo data demonstrate the importance of gcm autoregulation in NGB1-1A glia specification and the pivotal role of Hkb in the process.
Figure 6.
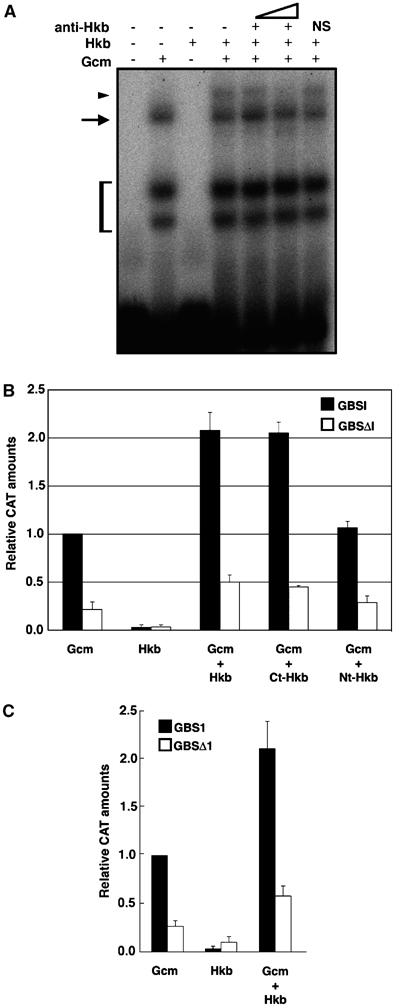
Gcm–Hkb synergistic activity. (A) Gel-shift assay showing DNA binding on a 30-mer containing GBSI. Arrow indicates binding of GST–Gcm N-terminal region (amino acids 1–261) to its target. Arrowhead indicates the additional band induced by incubation with both GST–Gcm N-terminal and GST–Hkb. Note that GST–Hkb does not, on its own, bind GBSI. Band shift induced by GST–Hkb is progressively removed by adding increasing amounts of anti-Hkb, but not by adding anti-Flag, used as nonspecific antibody (NS). Bracket indicates degraded GST–Gcm N-terminal products. (B) Cotransfection of reporter constructs containing either wild type (WT) or mutated GBSI (GBSI and ΔGBSI, see Figure 4) with pPAC-Gcm or pPAC-Hkb alone, or with pPAC-Gcm in combination with one of the Hkb-containing plasmids (pPAC-Hkb, pPAC-Ct-Hkb or pPAC-Nt-Hkb). (C) Cotransfection assays as in (B) but using reporter constructs containing either WT or mutated GBS1, a GBS that is three times more active than GBSI (Ragone et al, 2003; GBSC in Miller et al, 1998). CAT values obtained upon cotransfection with pPAC-Gcm and WT reporter were arbitrarily given a value of 1 and used for normalization. Each bar represents the average of at least three measurements, and error bars indicate standard error.
As hkb2 embryos do express gcm albeit at very low levels (Figures 1K and L), we asked whether this allele is a hypomorph by sequencing its cDNA. hkb2 carries a G → A mutation in the third nucleotide of the start codon. Moreover, no protein can be detected by Western blot on extracts from hkb2 embryos (Supplementary Figure 2). These data altogether indicate that hkb2 represents a null allele and that hkb is necessary to amplify gcm expression, but not to induce the first boost of transcription.
Thus, in the NGB1-1A lineage, hkb does not work as a transcription factor. Rather, it triggers gcm autoregulation via protein–protein interaction.
Hkb amplifies gcm expression in vivo
The above data suggest that Hkb controls NGB1-1A glia specification by mediating positive gcm autoregulation. If Hkb is sufficient to trigger this process, its overexpression should lead to excess of Hkb-dependent glia and to Gcm overexpression. We overexpressed Hkb throughout the CNS and analyzed flies carrying the UAS-hkb construct (Myat and Andrew, 2002), the sca-GAL4 driver and the enhancer-trap line P101 (Klambt and Goodman, 1991). This enhancer trap line specifically expresses β-galactosidase (β-gal) in SPG, including NGB1-1A-derived SPG (A- and B-SPG) (Figures 7A–A″). Hkb overexpression does induce the formation of additional Repo positive cells (Figure 7B′), most of which also express the P101 SPG marker (Figures 7B and B″). The presence of additional SPG is accompanied by ectopic gcm expression (compare Figure 8A with 8B). Moreover, gcm mRNA persists until stage 14 (Figure 8D), whereas in wild-type (WT) NGB1-1A, it fades away by the end of stage 12 (Figure 8C).
Figure 7.
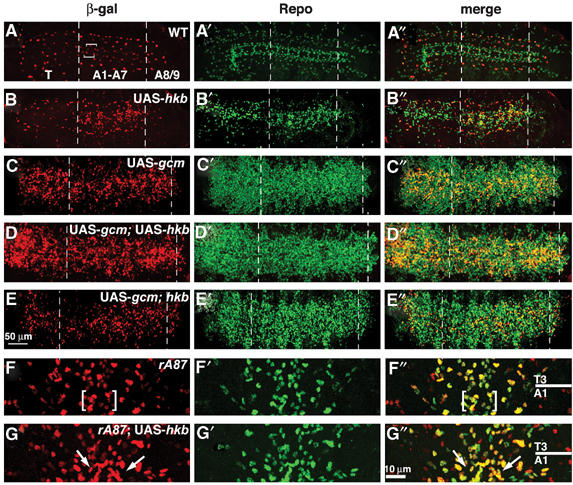
Role of Hkb and Gcm in NGB1-1A specification. (A–E″) Ventral views of stage 16 embryonic ventral cord carrying the P101 SPG marker; T3 and A1 indicate, respectively, third thoracic and first abdominal segments; anterior to the top and vertical line indicates the midline. β-gal (SPG), Repo double labeling. Left panels show β-gal, mid panels, Repo and right panels, merges. In all overexpression experiments, sca-GAL4 was used as a driver. (A–A″) labeling in wild-type (WT) embryo. Square brackets indicate A- and B-SPG (note that LV-SPG cannot be seen in this focal plane), arrowhead in (A) indicates lateral SPG, a cell that flanks A- and B-SPG cells, but is not derived from NGB1-1A lineage. (B–B″) labeling upon hkb overexpression induces additional SPG (B) and Repo (B′) labeling, close to the position at which A- and B-SPG are normally present. (C–C″) labeling upon combined gcm and hkb overexpression. Note the presence of additional SPG labeling in thoracic and abdominal segments (thick arrow). (D–D″) labeling upon gcm expression. (E–E″) labeling upon gcm expression in hkb embryo. (F–G″) Ventral views of stage 16 ventral cord of rA87/+ embryos. β-gal (rA87), Repo double labeling as above. (F–F″) labeling in a WT rA87/+ embryo. (G–G″) labeling in a sca-GAL4/rA87; UAS-hkb/+ embryo. Square brackets in (F and F″) indicate β-gal/Repo positive cells at the position of A- and B-SPG. Colocalization of additional Repo and β-gal labeling is indicated by arrows. Scale bar: 10 μm.
Figure 8.
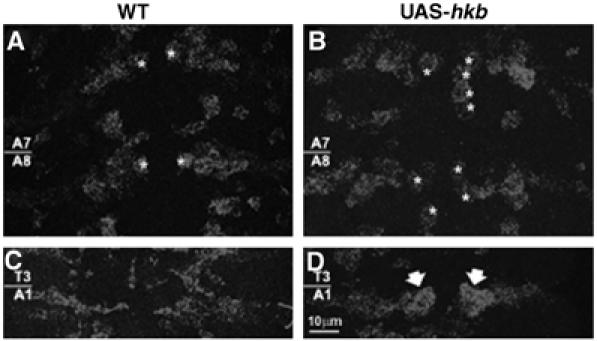
Hkb induces gcm expression in vivo. (A) gcm mRNA is expressed in one cell per hemisegment at stage 11 in a wild-type (WT) embryo (asterisks). (B) Several gcm expressing cells, indicated by white asterisks, are detectable in embryos overexpressing hkb (UAS-Hkb). sca-GAL4 driver was used for overexpression. Abdominal segments 7 and 8 (A7 and A8) are shown in (A) and (B). gcm expression profile at stage 14 in WT (C) and UAS-hkb (D) embryos. Thick arrows indicate persistent gcm expression in hkb overexpressing embryos. Scale bar: 10 μm.
To confirm the role of Hkb as a positive cofactor in gcm autoregulation, we analyzed rA87/sca-GAL4; UAS-hkb embryos. As in the rA87 enhancer trap line the lacZ gene is under the control of gcm promoter, β-gal expression can be used to trace autoregulation in vivo (Figures 7F–F″) (Miller et al, 1998). The additional Repo positive cells present in animals overexpressing hkb also express β-gal (Figures 7G–G″), meaning that Hkb positively acts on the gcm promoter and regulates its expression. In summary, the in vivo data show that hkb amplifies gcm levels by positive autoregulation and that this triggers NGB1-1A glia specification. The importance of gcm autoregulation is further confirmed by the observation that gcm overexpression in a WT background induces many more Repo positive cells than in a gcm mutant background (Figure 9).
Figure 9.
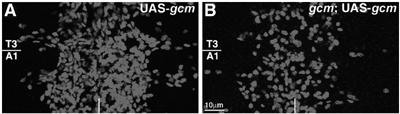
Glial differentiation requires gcm autoregulation. Ventral views of stage 16 embryos, Repo labeling upon gcm overexpression (sca-GAL4) in wild type (WT) (A) or gcm background (B). Symbols as in Figure 1. Scale bar: 10 μm.
The effects of hkb overexpression on gcm autoregulation and SPG specification prompted us to determine the relative contribution of gcm in this process. Indeed, co-overexpression of gcm and hkb induces many more Repo and SPG positive cells (Figures 7C–C″) compared to those observed upon hkb overexpression (Figures 7B–B″). Finally, overexpression of gcm alone is sufficient to induce SPG (Figures 7D–D″), even though less efficiently than in combination with hkb. Further confirming the importance of Gcm threshold levels, the presence or absence of endogenous hkb does not seem to modify the phenotype induced by Gcm overexpression (Figures 7D–D″ and E–E″, respectively), which over-rides the need for cell-specific autoregulation. In order to quantify the effect of gcm overexpression, the number of SPG was determined. Values obtained by counting 24 hemisegments per embryo (n=3) confirm that hkb, gcm cooverexpression leads to many more SPG (average: 820 nuclei/embryo) than gcm expression (average: 576 nuclei/embryo) and that hkb absence does not affect the phenotype induced by gcm overexpression (average: 549 nuclei/embryo). It is worth noting that, while differences can be observed with respect to SPG labeling, the three genotypes display a similar number of Repo positive cells, further confirming that hkb acts on glia specification.
Thus, hkb is necessary to sustain gcm expression in NGB1-1A lineage, which in turn induces SPG specification.
Discussion
We here show that Hkb controls glia specification by binding Gcm glia promoting factor and inducing high levels of gcm expression. Thus, cell-specific autoregulation of a fate determinant coordinates patterning and differentiation and is crucial for the establishment of cell diversity.
Importance of quantitative regulation during development
Autonomous and nonautonomous cues trigger specific cell fates in the nervous system and thereby guarantee its precise architecture. Morphogens elicit different fates depending on their concentration, a clear example being provided by Sonic hedgehog, the gradient of which specifies D/V positions within the neural tube (Poh et al, 2002). Autonomous cues, on the other hand, are known to trigger cell fates based on their cell-specific expression, emphasizing the importance of qualitative differences in the establishment of cell diversity (Bardin et al, 2004). Recent studies, however, call for a role of quantitative regulation even in the case of autonomous cues. Generation of neural precursors relies on progressive accumulation of proneural proteins, first in a group of cells called the proneural cluster and later on in one cell of the cluster, based on feedback loop interactions that control proneural protein levels in different cells (Skeath and Carroll, 1994; Baker, 2000; Ramain et al, 2000; zur Lage et al, 2004). Fate determinant levels are also important for glial differentiation: (i) the number of supernumerary glia depends on the amount of ectopic Gcm (Bernardoni et al, 1998); (ii) gcm RNA is unequally distributed in the dividing NGB, the presumptive neuroblast inheriting less RNA than the presumptive glioblast (Akiyama-Oda et al, 1999; Bernardoni et al, 1999; Ragone et al, 2001); (iii) gcm contains a PEST motif, characteristic of proteins at high turn over (Rogers et al, 1986), and an mRNA instability element (IE) (Shaw and Kamen, 1986) in the 3′ untranslated region, both of which are conserved throughout evolution (Hosoya et al, 1995; Kammerer et al, 1999; Kanemura et al, 1999; Tuerk et al, 2000; Wegner and Riethmacher, 2001; Hashemolhosseini et al, 2004). The present data show that gcm levels control not only the number but also the type of induced glia, demonstrating for the first time that autonomous cue quantitative regulation controls cell specification within a neural lineage.
Control of fate determinant levels has been shown to be mediated by cooperative pathways. Pannier prepatterning transcription factor interacts with proneural proteins and activates transcription through its DNA target sequences in order to modulate proneural gene expression (Ramain et al, 2000). Long-range-mediated interactions facilitate such cooperativity, as Chip (Morcillo et al, 1997) interaction with both Pannier and Achaete enhances Achaete autoregulation (Ramain et al, 2000). Also, autoregulation of Atonal (Ato) proneural protein, which is necessary for recruiting chordotonal sensory organ precursors (Jarman et al, 1993), depends on the cooperative activity of Ato and Pointed P1 transcription factors, which bind E box and ETS motifs, respectively (zur Lage et al, 2004). Strikingly, gcm autoregulation in NGB1-1A does not depend on cooperative activation of two transcriptional pathways. Rather, the presence of the GBS is sufficient in vitro and in vivo for cell-specific gcm autoregulation, thus pointing to a novel molecular strategy controlling fate determinant levels.
The role of Hkb is to sustain gcm-dependent transcription by acting on Gcm binding and transactivation potential. This is also supported by three observations: (i) Hkb works on different GBSs; (ii) mutagenized GBSI is active, although at low levels, in the presence of Hkb and Gcm, but not in the presence of Gcm alone; (iii) gcm overexpression does produce ectopic SPG in an hkb context, as this overcomes the need for autoregulation. In the establishment of terminal patterning in the fly embryo, hkb works as a repressor via interaction of its Nt region with Groucho protein (Goldstein et al, 1999). This region, however, is neither relevant for Hkb binding to Gcm nor for activating gcm autoregulation. Thus, hkb works either as a repressor or as a coactivator by interacting with different proteins.
Cell differentiation and specification in the nervous system
Glia specificity (e.g., longitudinal versus SPG) could rely on genes that are activated by a pathway independent of Gcm and impose specification on postmitotic cells otherwise displaying a default ‘pan-glia' fate. Specification of postmitotic cells does also take place during the development of neurons, where it refines/maintains early decisions taken in precursor cells and allows for further diversification (Allan and Thor, 2003, 2005). While establishing the possible need for postmitotic specification awaits the identification of novel glial markers, present data demonstrate the importance of cues working in glial precursors.
Integrating cell differentiation and specification via Gcm–Hkb interaction allows a single fate determinant to generate different types of glia. It will be interesting to determine whether Hkb also acts on gcm targets, as it is known that some of these targets are pan-glial (Repo: Campbell et al, 1994; Xiong et al, 1994; Halter et al, 1995), whereas others are lineage specific (Loco: Granderath et al, 2000). It will also be interesting to determine whether gcm autoregulation mediated by cell-specific factors is necessary in other glial lineages.
Our data show that NGB1-1A glia specification requires two equally important regulatory steps. The first one is gcm independent and results in low levels of gcm expression, whereas the second one is gcm and hkb dependent and induces high levels of gcm. Previous data show that hkb expression is induced by columnar genes, which control D/V regionalization in the nervous system (Chu et al, 1998; McDonald et al, 1998; Mellerick and Modica, 2002). Thus, cell-specific factors such as Hkb provide an intermediate step between patterning genes and fate determinants, thereby triggering the identity of neural precursors.
Supernumerary P101 positive cells induced by Hkb overexpression are all located close to endogenous SPG. The central and abdominal position of supernumerary SPG suggests that cells within the NGB1-1A lineage are ‘competent' to express the SPG fate. Furthermore, hkb gliogenic activity is also temporally restricted, since the first division is never affected and always produces aCC/pCC sibling neurons (Supplementary Figure 3). These data call for additional factors regulating gcm expression and SPG specification and, indeed, homeotic as well as temporal genes are known to control the NGB1A-A lineage (Udolph et al, 1993; Prokop and Technau, 1994; Isshiki et al, 2001). Thus, a grid of positional cues along spatial and temporal axes works through gcm, thereby triggering differentiation of the appropriate type of glia. Understanding the interplay of these cues will be the matter of further studies; however, our data already demonstrate that cofactor-mediated quantitative regulation plays a pivotal role in cell specification. Such fine-tuning and accurate orchestration of events highlights the complexity underlying nervous system differentiation. Integrating qualitative and quantitative regulation via cell-specific autoregulation likely applies to other developmental processes in which a single fate determinant triggers different phenotypes.
Materials and methods
Stocks
WT strain was Sevelen. gcm26 (null allele) and hkb2 (Weigel et al, 1990) were used as mutant strains. Homozygous mutant embryos were identified by using blue balancers (β-gal labeling as in Ashburner, 1989). scabrous-GAL4 (sca-GAL4) was used to express UAS-gcm (Bernardoni et al, 1998), UAS-hkb (Myat and Andrew, 2002) and UAS-Ubx (Castelli-Gair et al, 1994). rA87 line is described in Vincent et al (1996). P101 enhancer trap line (Klambt and Goodman, 1991) was used as SPG marker. [w; gcm26/CyO twi-lacZ; P(2kb-gcm,w+)] and [w; P(2kb-gcm,w+); hkb2/TM3, Ser, twi-lacZ] lines carry the WT transgene in gcm or hkb background, respectively. [w; gcm26/CyO twi-lacZ; P(2kbΔHBS-gcm,w+)] and [w; gcm26/CyO twi-lacZ; P(2kbΔGBSI-gcm,w+)] carry the mutant transgenes in a gcm background.
In situ hybridization and immunolabeling
Embryo preparation, antibody incubation and in situ hybridization were performed as in Bernardoni et al (1997). Digoxigenin-labeled gcm and Fluorescein-labeled hkb riboprobes were obtained by using full-length cDNAs. Embryos were mounted in Vectashield medium (Vector). The following primary antibodies were used: rabbit anti-Repo (1:1000) (A Travers), mouse anti-Repo (1:100) (DHSB), rabbit anti-PH3 (1:10000) (Upstate Biotechnology), mouse anti-digoxigenin (1:100) (Boehringer), rabbit anti-fluorescein (1:1000) (Molecular Probes), mouse anti-β-gal (1:100) (DSHB), rabbit anti-β-gal (1:500) (Cappel). Secondary antibodies coupled to Cy3 and FITC (Jackson) were used at 1:400. Chromatin labeling was obtained by using DAPI at 100 ng/ml in PBS-0.3% Triton X-100. Preparations were analyzed by confocal microscopy (DMRE, Leica).
Cell transfection and CAT ELISA assay
Reporter vector pBLCAT5-GBSI and pBLCAT5-GBS1 as well as vectors pPAC5C (which we refer to as pPAC, C Thummel) and pPAC-Gcm are described in Miller et al (1998). pBLCAT5-ΔGBSI and pBLCAT5-ΔGBS1 were obtained by double-stranded mutagenesis (Clontech) on pBLCAT5-GBSI and pBLCAT5-GBS1, respectively, using the following primers:
- ΔGBSI:
5′-GGATTCTAATGTTTCCCTTAAAGGATTC-3′;
5′-GGATTCTGCAAGGGAAACATCTGGATTC-3′.
- ΔGBS1:
5′-GGATTCTGCAAGGGAAACATCTGGATTC-3′.
ΔGBSI: 5′-GGATTCTAATGTTTCCCTTAAAGGATTC-3′;
ΔGBS1: 5′-GGATTCTGCAAGGGAAACATCTGGATTC-3′.
pPAC-Hkb, pPAC-Nt-Hkb and pPAC-Ct-Hkb were obtained by cloning the full-length, the first 471nt or the last 411nt of hkb cDNA in the pPAC vector, respectively. pPAC-LacZ was obtained by cloning the entire lacZ cDNA in pPAC vector. Transient transfection of Drosophila S2 line (Schneider, 1972) was performed using effectene (Qiagen), according to the manufacturer's instructions, using 1.15 μg of DNA containing the following: 100 ng of pPAC-LacZ, 50 ng of reporter DNA, 1 μg of expression vectors. Cells were harvested 48 h after transfection and normalized for β-gal activity. CAT levels were determined using the CAT ELISA kit (Boehringer).
In vitro GST pull-down assays
An EcoRI fragment containing the hkb full-length cDNA was cloned into the pGEX4T3 (Pharmacia) to produce a GST–Hkb fusion protein of 50 kDa. GST–Hkb was expressed in the protease-deficient E. coli strain BL21 and purified using GST-conjugated Sepharose 4B (Pharmacia), according to the manufacturer's instructions. The GST–Nt-Hkb and GST–Ct-Hkb were produced in the same manner by cloning the first 471nt or the last 411nt of hkb cDNA, corresponding to 157 amino acids of the amino terminal part or 137 amino acids of the carboxy terminal part of Hkb, respectively. Luciferase and Gcm (full length, Gcm 1–421 and Gcm 261–421) proteins were produced by in vitro transcription–translation in rabbit reticulocyte lysate in the presence of methionine, according to manufacturer's instructions (Promega, Madison, WI). Pull-down assays were performed using 1 μg of fusion protein, bound to 50 μl beads and preincubated with 1 ml of binding buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EGTA, 1% Triton X-100, 1 mM PMSF). [35S]Gcm or Luciferase proteins were added to each preincubation mix and binding reactions were carried out overnight at 4°C. Beads were washed several times with binding buffer, boiled for 5 min in sample buffer and aliquots were examined by electrophoresis. The amount of retained proteins was detected by autoradiography.
DNA-binding assay
An EcoRI fragment containing the first 261 amino acids of Gcm was generated by PCR and cloned into pGEX4T3 (Pharmacia) to produce GST–Gcm Nt fusion protein, which was used in gel-shift assays (Miller et al, 1998). Same 30-mer containing the GBSI as in Miller et al (1998).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank A Travers, D Andrew, J Castelli-Gair, J Urban, MA Krasnow, the Bloomington Stock Center and the DSHB for flies, antibodies and plasmids. We also thank M Boeglin, JL Vonesch and D Hentsch for help with confocal microscopy, imaging and data processing. We thank all group members for helpful comments on the manuscript and C Diebold for excellent technical assistance. Thanks to N Arbogast for keeping fly stocks. This work was supported by EEC (QLG3-CT-2000-01224), INSERM, CNRS, the Hôpital Universitaire de Strasbourg, Ministère de la Recherche (ACI Développement) and by ARC. RD was supported by European Leucodystrophies Association (ELA), Ligue Nationale contre le Cancer (LIGUE) and Association pour la Recherche de la Sclerose en Plaque fellowships. MK was supported by LIGUE, BDI (CNRS) and ELA fellowships, and SS by LIGUE and EEC fellowhips. LS by LIGUE and Association Française contre les Myopathies fellowships. CJ was supported by an MRT fellowship.
References
- Akiyama-Oda Y, Hosoya T, Hotta Y (1998) Alteration of cell fate by ectopic expression of Drosophila glial cells missing in non-neural cells. Dev Genes Evol 208: 578–585 [DOI] [PubMed] [Google Scholar]
- Akiyama-Oda Y, Hosoya T, Hotta Y (1999) Asymmetric cell division of thoracic neuroblast 6–4 to bifurcate glial and neuronal lineage in Drosophila. Development 126: 1967–1974 [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Hosoya T, Poole AM, Hotta Y (1996) The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc Natl Acad Sci USA 93: 14912–14916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan DW, Park D, St Pierre SE, Taghert PH, Thor S (2005) Regulators acting in combinatorial codes also act independently in single differentiating neurons. Neuron 45: 689–700 [DOI] [PubMed] [Google Scholar]
- Allan DW, Thor S (2003) Together at last: bHLH and LIM-HD regulators cooperate to specify motor neurons. Neuron 38: 675–677 [DOI] [PubMed] [Google Scholar]
- Ashburner M (1989) Drosophila: A laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Baker NE (2000) Notch signaling in the nervous system. Pieces still missing from the puzzle. Bioessays 22: 264–273 [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Le Borgne R, Schweisguth F (2004) Asymmetric localization and function of cell-fate determinants: a fly's view. Curr Opin Neurobiol 14: 6–14 [DOI] [PubMed] [Google Scholar]
- Bernardoni R, Kammerer M, Vonesch JL, Giangrande A (1999) Gliogenesis depends on glide/gcm through asymmetric division of neuroglioblasts. Dev Biol 216: 265–275 [DOI] [PubMed] [Google Scholar]
- Bernardoni R, Miller AA, Giangrande A (1998) Glial differentiation does not require a neural ground state. Development 125: 3189–3200 [DOI] [PubMed] [Google Scholar]
- Bernardoni R, Vivancos V, Giangrande A (1997) glide/gcm is expressed and required in the scavenger cell lineage. Dev Biol 191: 118–130 [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F (2002) Proneural genes and the specification of neural cell types. Nat Rev Neurosci 3: 517–530 [DOI] [PubMed] [Google Scholar]
- Bossing T, Technau GM, Doe CQ (1996a) huckebein is required for glial development and axon pathfinding in the neuroblast 1-1 and neuroblast 2-2 lineages in the Drosophila central nervous system. Mech Dev 55: 53–64 [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM (1996b) The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol 179: 41–64 [DOI] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ (1995) New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Dev 53: 393–402 [DOI] [PubMed] [Google Scholar]
- Bronner G, Chu-LaGraff Q, Doe CQ, Cohen B, Weigel D, Taubert H, Jackle H (1994) Sp1/egr-like zinc-finger protein required for endoderm specification and germ-layer formation in Drosophila. Nature 369: 664–668 [DOI] [PubMed] [Google Scholar]
- Bronner G, Jackle H (1991) Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech Dev 35: 205–211 [DOI] [PubMed] [Google Scholar]
- Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A (1994) RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development 120: 2957–2966 [DOI] [PubMed] [Google Scholar]
- Castelli-Gair J, Greig S, Micklem G, Akam M (1994) Dissecting the temporal requirements for homeotic gene function. Development 120: 1983–1995 [DOI] [PubMed] [Google Scholar]
- Chu H, Parras C, White K, Jimenez F (1998) Formation and specification of ventral neuroblasts is controlled by vnd in Drosophila neurogenesis. Genes Dev 12: 3613–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-LaGraff Q, Schmid A, Leidel J, Bronner G, Jackle H, Doe CQ (1995) huckebein specifies aspects of CNS precursor identity required for motoneuron axon pathfinding. Neuron 15: 1041–1051 [DOI] [PubMed] [Google Scholar]
- Goldstein RE, Jimenez G, Cook O, Gur D, Paroush Z (1999) Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development 126: 3747–3755 [DOI] [PubMed] [Google Scholar]
- Granderath S, Bunse I, Klambt C (2000) gcm and pointed synergistically control glial transcription of the Drosophila gene loco. Mech Dev 91: 197–208 [DOI] [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM (1995) The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development 121: 317–332 [DOI] [PubMed] [Google Scholar]
- Hashemolhosseini S, Schmidt K, Kilian K, Rodriguez E, Wegner M (2004) Conservation and variation of structure and function in a newly identified GCM homolog from chicken. J Mol Biol 336: 441–451 [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y (1995) Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 82: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ (2001) Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106: 511–521 [DOI] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau GM (1995) Distribution, classification and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Roux's Arch Dev biol 204: 284–307 [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN (1993) atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73: 1307–1321 [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS (1995) glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell 82: 1013–1023 [DOI] [PubMed] [Google Scholar]
- Kammerer M, Pirola B, Giglio S, Giangrande A (1999) GCMB, a second human homolog of the fly glide/gcm gene. Cytogenet Cell Genet 84: 43–47 [DOI] [PubMed] [Google Scholar]
- Kanemura Y, Hiraga S, Arita N, Ohnishi T, Izumoto S, Mori K, Matsumura H, Yamasaki M, Fushiki S, Yoshimine T (1999) Isolation and expression analysis of a novel human homologue of the Drosophila glial cells missing (gcm) gene. FEBS Lett 442: 151–156 [DOI] [PubMed] [Google Scholar]
- Klambt C, Goodman CS (1991) The diversity and pattern of glia during axon pathway formation in the Drosophila embryo. Glia 4: 205–213 [DOI] [PubMed] [Google Scholar]
- Kuhnlein RP, Bronner G, Taubert H, Schuh R (1997) Regulation of Drosophila spalt gene expression. Mech Dev 66: 107–118 [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL (2003) Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38: 731–745 [DOI] [PubMed] [Google Scholar]
- McDonald JA, Holbrook S, Isshiki T, Weiss J, Doe CQ, Mellerick DM (1998) Dorsoventral patterning in the Drosophila central nervous system: the vnd homeobox gene specifies ventral column identity. Genes Dev 12: 3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerick DM, Modica V (2002) Regulated vnd expression is required for both neural and glial specification in Drosophila. J Neurobiol 50: 118–136 [DOI] [PubMed] [Google Scholar]
- Miller AA, Bernardoni R, Giangrande A (1998) Positive autoregulation of the glial promoting factor glide/gcm. EMBO J 17: 6316–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D (1997) Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev 11: 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ (2002) Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell 111: 879–891 [DOI] [PubMed] [Google Scholar]
- Poh A, Karunaratne A, Kolle G, Huang N, Smith E, Starkey J, Wen D, Wilson I, Yamada T, Hargrave M (2002) Patterning of the vertebrate ventral spinal cord. Int J Dev Biol 46: 597–608 [PubMed] [Google Scholar]
- Powell LM, Zur Lage PI, Prentice DR, Senthinathan B, Jarman AP (2004) The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol 24: 9517–9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A, Technau GM (1994) Early tagma-specific commitment of Drosophila CNS progenitor NB1-1. Development 120: 2567–2578 [DOI] [PubMed] [Google Scholar]
- Ragone G, Bernardoni R, Giangrande A (2001) A novel mode of asymmetric division identifies the fly neuroglioblast 6-4T. Dev Biol 235: 74–85 [DOI] [PubMed] [Google Scholar]
- Ragone G, Van De Bor V, Sorrentino S, Kammerer M, Galy A, Schenck A, Bernardoni R, Miller AA, Roy N, Giangrande A (2003) Transcriptional regulation of glial cell specification. Dev Biol 255: 138–150 [DOI] [PubMed] [Google Scholar]
- Ramain P, Khechumian R, Khechumian K, Arbogast N, Ackermann C, Heitzler P (2000) Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol Cell 6: 781–790 [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234: 364–368 [DOI] [PubMed] [Google Scholar]
- Rowitch DH (2004) Glial specification in the vertebrate neural tube. Nat Rev Neurosci 5: 409–419 [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM (1997) The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol 189: 186–204 [DOI] [PubMed] [Google Scholar]
- Schneider I (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 27: 353–365 [PubMed] [Google Scholar]
- Schreiber J, Sock E, Wegner M (1997) The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain. Proc Natl Acad Sci USA 94: 4739–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Kamen R (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46: 659–667 [DOI] [PubMed] [Google Scholar]
- Skeath JB (1999) At the nexus between pattern formation and cell-type specification: the generation of individual neuroblast fates in the Drosophila embryonic central nervous system. Bioessays 21: 922–931 [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB (1994) The achaete-scute complex: generation of cellular pattern and fate within the Drosophila nervous system. FASEB J 8: 714–721 [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S (2003) Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol 13: 8–15 [DOI] [PubMed] [Google Scholar]
- Soustelle L, Giangrande G (2005) Early Embryonic Development: Neurogenesis (CNS). In Comprehensive Molecular Insect Science Vol 1, Lawrence I. Gilbert, Kostas Iatrou, Sarjeet Gill (eds) pp 343–378. Oxford: Elsevier Press [Google Scholar]
- Tuerk EE, Schreiber J, Wegner M (2000) Protein stability and domain topology determine the transcriptional activity of the mammalian glial cells missing homolog, GCMb. J Biol Chem 275: 4774–4782 [DOI] [PubMed] [Google Scholar]
- Udolph G, Prokop A, Bossing T, Technau GM (1993) A common precursor for glia and neurons in the embryonic CNS of Drosophila gives rise to segment-specific lineage variants. Development 118: 765–775 [DOI] [PubMed] [Google Scholar]
- Udolph G, Rath P, Chia W (2001) A requirement for Notch in the genesis of a subset of glial cells in the Drosophila embryonic central nervous system which arise through asymmetric divisions. Development 128: 1457–1466 [DOI] [PubMed] [Google Scholar]
- Van De Bor V, Giangrande A (2002) glide/gcm: at the crossroads between neurons and glia. Curr Opin Genet Dev 12: 465–472 [DOI] [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A (1996) Glide directs glial fate commitment and cell fate switch between neurones and glia. Development 122: 131–139 [DOI] [PubMed] [Google Scholar]
- Wegner M, Riethmacher D (2001) Chronicles of a switch hunt: gcm genes in development. Trends Genet 17: 286–290 [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Klingler M, Jackle H (1990) Two gap genes mediate maternal terminal pattern information in Drosophila. Science 248: 495–498 [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C (1994) repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev 8: 981–994 [DOI] [PubMed] [Google Scholar]
- zur Lage PI, Powell LM, Prentice DR, McLaughlin P, Jarman AP (2004) EGF receptor signaling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev Cell 7: 687–696 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


