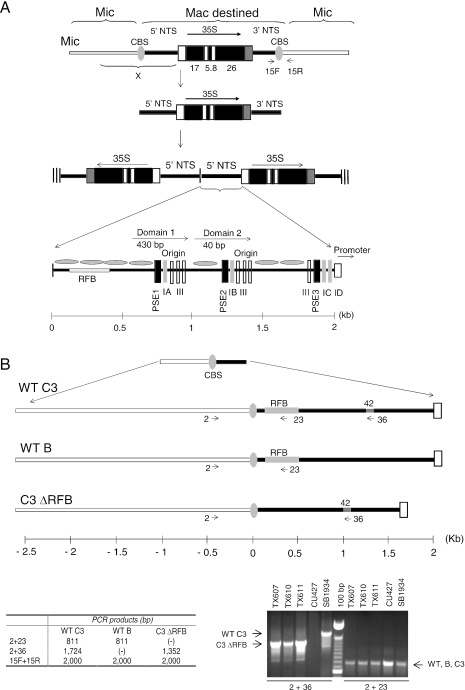
Figure 1.
rDNA processing pathway. (A) Organization of the rDNA in the micro- and macronucleus. The gene encoding the 35S precursor RNA for 17S, 5.8S and 26S ribosomal RNAs, exists as a single ‘integrated’ copy in micronuclear chromosome 1. Macronuclear-destined rDNA sequences are excised from this chromosome in the newly developing macronucleus by cleavage at flanking Cbs elements. The released rDNA monomer is subsequently rearranged into a giant head-to-head ‘palindrome’ with two inverted copies of the 5′ NTS at the center and telomeres at each terminus (hatched vertical lines). The 1.9 kb 5′ NTS (bottom panel) contains positioned nucleosomes that bracket the rRNA promoter and replication initiation sites, the later of which reside within the tandemly reiterated Domain 1 and 2 (D1, D2) regions. These nucleosome-free regions contain conserved cis-acting determinants that regulate replication initiation and fork progression, designated PSEs (PSE1, 2 and 3; vertical black boxes) and type 1 elements (1A–1D; vertical grey boxes). The approximate position of the developmentally regulated RFB is shown. (B) Polymorphisms for micronuclear rDNA allele analysis. Top and lower left panels: enlargement of the micronuclear chromosome 1 region designated as ‘X’ in panel A (top line), showing the RFB region and a 42 bp sequence that is present in wild-type C3 and C3ΔRFB alleles, but is absent in the naturally occurring B rDNA allele. The positions of PCR primers that anneal to the respective rDNA species are shown (arrows), along with the predicted sizes of PCR products (table). Primers 15F and 15R span the 3′ Cbs element and amplify the micronuclear rDNA species for all three alleles. Lower right panel: PCR analysis of micronuclear rDNA in wild-type C3 (SB1934) and wild-type B (CU427) rDNA strains, and heterozygous micronuclear transformants harboring the C3ΔRFB and B rDNA alleles (TX607, TX610 and TX611).