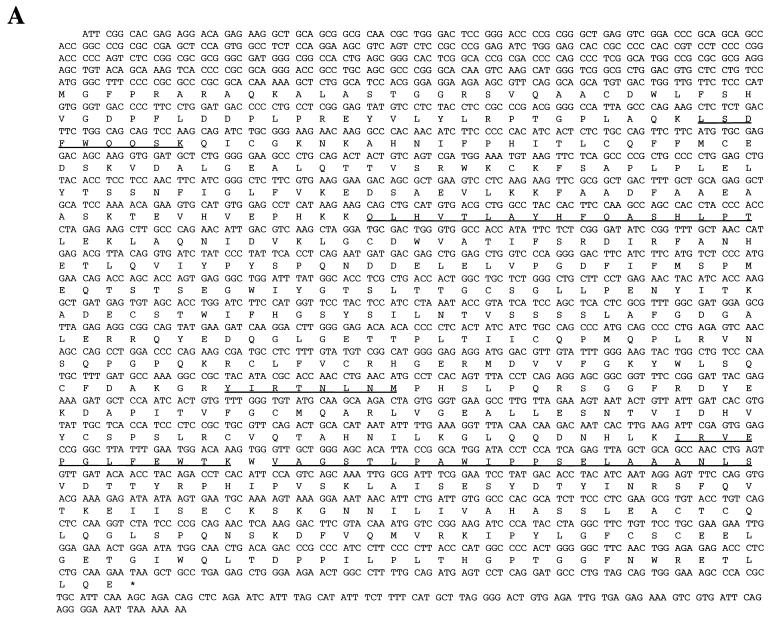
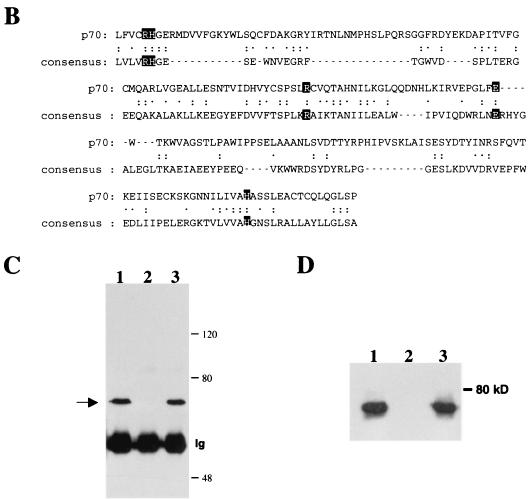
FIG. 2.
The p70 gene. (A) Nucleotide sequence and predicted amino acid sequence of p70 open reading frame. Underlined amino acids indicate peptides obtained from microsequencing-purified p70 protein, as described in Materials and Methods. (B) Alignment of C-terminal region of p70 with a consensus sequence built by the Protein Family (Pfam) database for members of the phosphoglycerate mutase family (1). The two arginines, two histidines, and glutamic acid predicted to be involved in catalytic activity are highlighted. Identical residues are indicated with two dots; chemically similar residues are indicated with one dot. (C) Peptide competition analysis. Proteins precipitated from lysates of DA3 cells by anti-p70 peptide antibody were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was probed with anti-p70 peptide antibody. Prior to immunoprecipitation, lysates were incubated for 15 min with the following: lane 1, no peptide; lane 2, antigenic peptide; and lane 3, unrelated peptide. Number of kilodaltons is given on right. (D) Interaction of p70 with phosphorylated Jak2 Y966 peptide. DA3 cytoplasmic lysates were subjected to immunoprecipitation with anti-p70 peptide antibodies (lane 1) or affinity pulldown assays utilizing nonphosphorylated Y966 peptide conjugated to Sepharose (lane 2) or phosphorylated Y966 peptide conjugated to Sepharose (lane 3). Blotting antibody was anti-p70 peptide antibody.