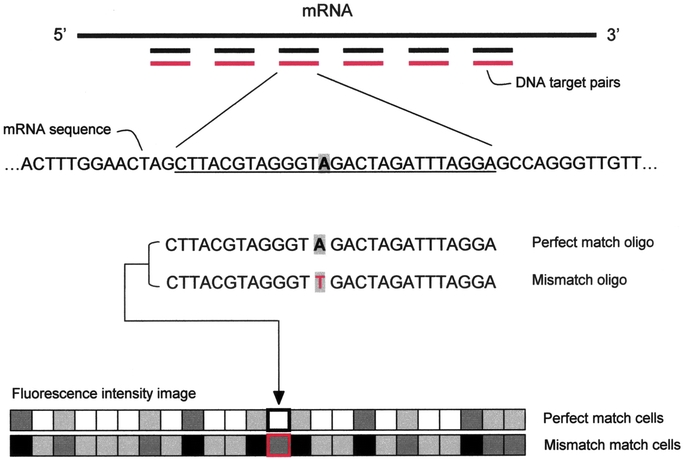
FIGURE 2. Oligonucleotide array scheme. The key point for this DNA array platform is the targeted design of probe sets. Using as little as 200 to 300 bases of gene, cDNA, or EST sequence, independent 25-mer oligonucleotides are selected to serve as unique, sequence-specific detectors. The arrays are designed in silico, and as a result, it is not necessary to prepare, verify, quantitate, and catalog a large number of cDNAs, polymerase chain reaction products, and clones, and there is no risk of a misidentified tube, clone, cDNA, or spot. Crucial for this approach is the use of target redundancy, which is not meant as the deposition of the same piece of DNA in multiple locations on an array, but rather the use of multiple oligonucleotides of different sequences designed to hybridize to different regions of the same RNA. The use of multiple independent detectors for the same molecule greatly improves signal-to-noise ratios, improves the accuracy of RNA quantitation, reduces the effects of crosshybridization, and drastically decreases the rate of false-positives. For each gene monitored, 32 oligonucleotides are synthesized, using photolithography, directly onto the chip. Oligonucleotides are arranged as 16 pairs: each pair includes a perfect match 25mer, which is an exact complement to the gene sequence, and a control oligonucleotide, which differs from the perfect match oligo at the 13th base. The reported hybridization intensity is a composite of the 16 perfect match–mismatch differences per gene. This redundancy considerably increases the statistical power of the technology and data can be analyzed using standard statistical techniques18 or by taking advantage of the changes in relative hybridization between perfect match and mismatch oligos to define genes as absent, present, increased, or decreased according to a set of heuristic rules.16 The mismatch probes act as specificity controls that allow the direct subtraction of both background and crosshybridization signals, and allow discrimination between “real” signals and those resulting from nonspecific or semispecific hybridization, which are more likely to occur with single-spot strategy DNA arrays (eg, cDNA array platform). In the presence of even low concentrations of RNA, hybridization to the perfect match/mismatch pairs produces recognizable and quantitative fluorescent patterns. The strength of these patterns directly relates to the concentration of the RNA molecules in the complex sample (even without a competitive hybridization or 2-color comparison).
