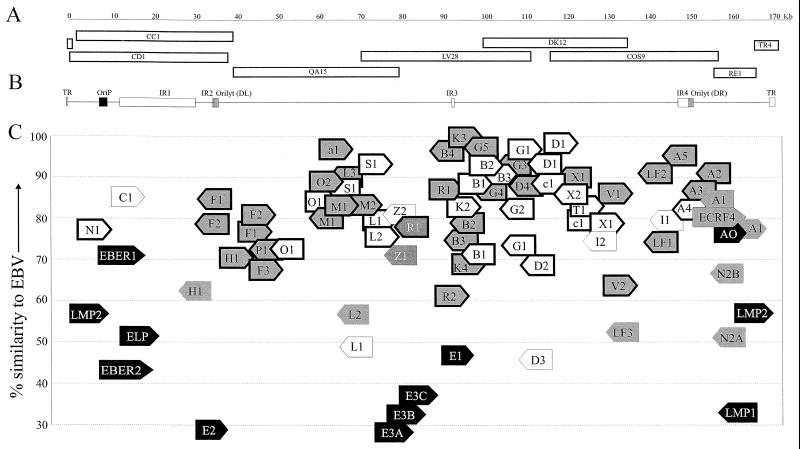
FIG. 1.
Rhesus LCV genome, ORFs, and homology with EBV ORFs. (A) Overlapping cosmid and plasmid DNA clones used to sequence the rhesus LCV genome. Cosmids were identified from the library by hybridization with the EBV BamHI C (CC1, CD1), BamHI Q (QA15), BamHI L (LV28), BamHI D (DK12), and BamHI A (cos9) DNA fragments. RE1 and TR4 are EcoRI and BamHI fragments, respectively, cloned from Hirt DNA. The nucleotide coordinates for each viral DNA clone are as follows: CD1 (140 to 38,206), CC1 (1,785 to 39,553), QA15 (39,641 to 79,990), LV28 (70,760 to 111,969), DK12 (100,417 to 135,417), Cos9 (116,690 to 157,414), RE1 (156,359 to 166,542), and TR4 (166,175 to 783). An 88-bp gap between the CC1 and QA15 cosmid clones was deduced from four PCR clones amplified from rhesus LCV-infected cell DNA using primers from the CC1 and QA15 sequence. (B) Organization of the rhesus LCV genome. Homologues for the EBV lytic and latent origins of replication (ori-p; 7,511 to 9,357), ori-lyt DL (34,141 to 35,138), ori-lyt DR (138,080 to 139,080), major repeat regions IR1 (12,240 to 29,750), IR2 (33,674 to 34,047), IR3 (89,780 to 90,460), and IR4 (135,263 to 137,761), and terminal repeats (TR; 167,326 to 171,106) are identified in the rhesus LCV genome as shown. (C) Rhesus LCV ORFs and amino acid homology with EBV ORFs. The percent amino acid similarity is shown on the y axis. Latent, immediate-early, early, and late lytic ORFs are in black, dark grey, light grey, and white, respectively. Latent infection genes are identified by name (LMPs, EBERs, EBNAs [E], and BARF0 [A0]). Each lytic infection ORF is identified using the EBV nomenclature for BamHI ORFs. The orientations of the ORFs are shown by the direction of the arrow (i.e., right or left). The EBV BamHI fragment is indicated by the letter within the arrow, and the number of the ORF in the EBV BamHI fragment is given last, e.g., the rhesus LCV BCRF1 homologue is indicated by the rightward C1 arrow with approximately 85% amino acid similarity. (The ECRF4 ORF is the only exception to these abbreviations.) ORFs common to other herpesviruses are shown with a bold outline. The initiator codon for each ORF is positioned accurately, but the ORF size is not drawn to scale.