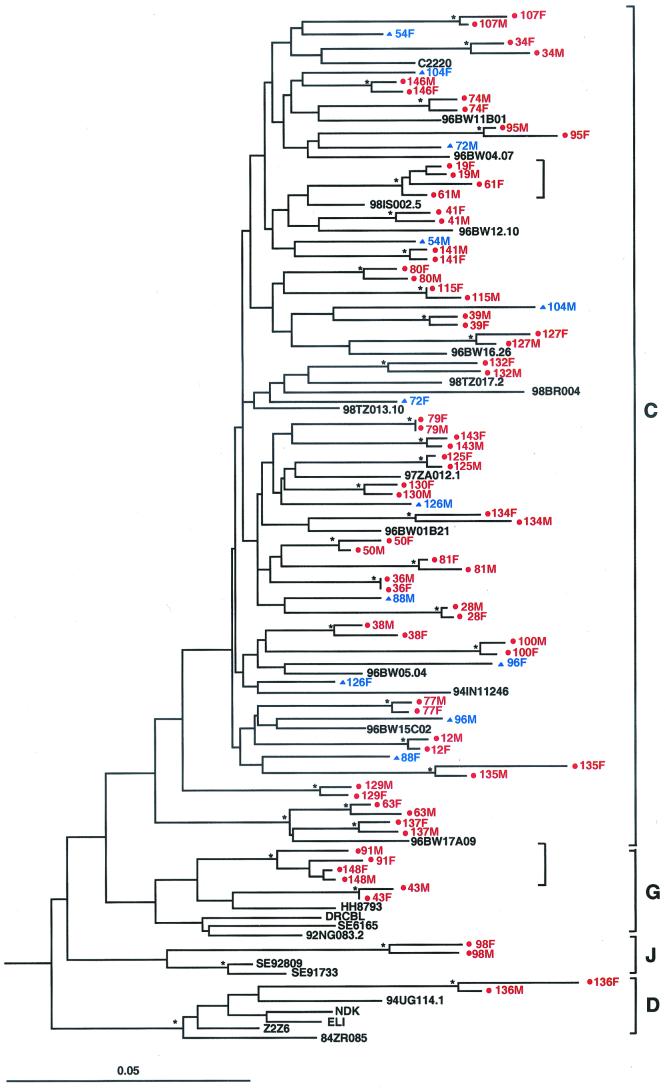
FIG. 3.
Molecular linkage analysis for a subset of putative HIV-1 transmission pairs. A phylogenetic tree was constructed from partial gp41 sequences (consensus length, 276 bp) by using the neighbor-joining method (34) and the Kimura two-parameter model (22). Horizontal branch lengths are drawn to scale (the scale bar represents 0.05 nucleotide substitutions per site); vertical separation is for clarity only. Asterisks indicate bootstrap values in which the cluster to the right is supported in >80% replicates (out of 1,000). Newly derived sequences from 42 transmission pairs (84 individuals) are shown, along with 26 reference sequences from the Los Alamos Sequence Database (http://hiv-web.lanl.gov/HTML/alignments.html). Viruses from 36 couples are closely related to one another and cluster together with significant bootstrap values, indicating that they are epidemiologically linked (highlighted in red and denoted by dots). Viruses from six couples do not cluster together and exhibit a range of within-couple diversity that is similar to that of the reference sequences (highlighted in blue and denoted by triangles), indicating that they are epidemiologically unlinked. Two small brackets denote viral subclusters, each involving viruses from two sets of couples (see text for details). Brackets on the far right indicate major group M sequence subtypes.