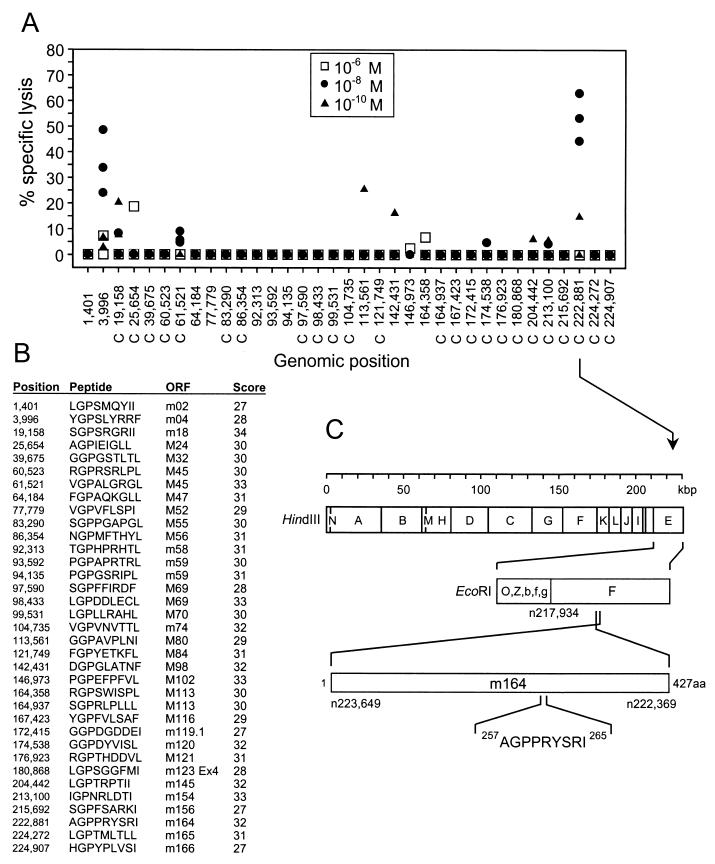
FIG. 1.
Identification of an antigenic peptide in ORF m164 of mCMV strain Smith. (A) A search for the Dd binding motif xGPxxxxx[L, I, F] was performed for all ORFs of the full-length genomic sequence of mCMV strain Smith (GenBank accession no. MCU68299), and the corresponding synthetic nonapeptides were used at the indicated molar concentrations for the generation of short-term microculture CTLLs. Data represent the cytolytic activity of individual microcultures tested on P815 target cells that were pulsed with the corresponding peptides at the corresponding concentrations. The genomic positions of the nonapeptide-coding sequences are given by the positions of the first nucleotides (n) according to the listing by Rawlinson et al. (43). C, complementary strand. (B) List of nonameric Dd-binding motifs. ORFs of mCMV that are sequence homologs of hCMV ORFs are indicated by capital M. Scores for Dd binding strengths were provided by Stefan Stevanovic (Institute for Cell Biology, Department of Immunology, University of Tuebingen, Tuebingen, Germany). (C) HindIII and EcoRI genetic map locations of ORF m164 peptide C 222881 257AGPPRYSRI265 drawn to scale. Amino acid positions refer to the deduced protein sequence according to Rawlinson et al. (43).