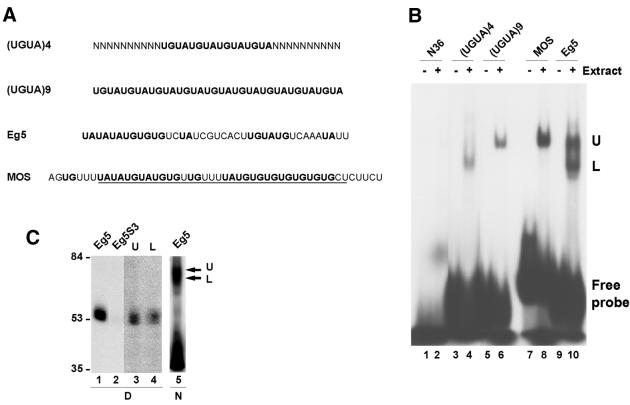
Figure 2.
Analysis of the EDEN-BP/UGUA repeat complex. (A) Sequence of the RNA molecules. U/purine dinucleotides are in bold. The minimal c-mos EDEN that is efficient in deadenylation is underlined. (B) RNA gel shift analysis. The indicated radiolabelled RNAs were incubated with (+) or without (–) 4 h post-fertilisation embryo extract. The complexes formed were analysed by native PAGE. The position of the free probes are indicated as well as the position of the two complexes formed: Upper (U) and Lower (L). (C) Coupled UV-induced label transfer and RNA gel shift. Lanes 1 and 2, UV-induced label transfer analysis of Eg5 and the mutant Eg5S3 RNAs incubated in embryo extracts. Lanes 3 and 4, gel slabs corresponding to the positions of arrows U and L in lane 5 were excised, treated with RNase A and deposited as indicated in the wells of a denaturing SDS–polyacrylamide gel. After electrophoresis the gel was dried and the radiolabelled proteins revealed by autoradiography. Lane 5, The Eg5 RNA was incubated in embryo extracts and UV irradiated prior to separation of the protein–RNA complexes on a native polyacrylamide gel and autoradiography of the unfixed wet gel. At the bottom of the figure, D indicates denaturing gel and N indicates native gel.