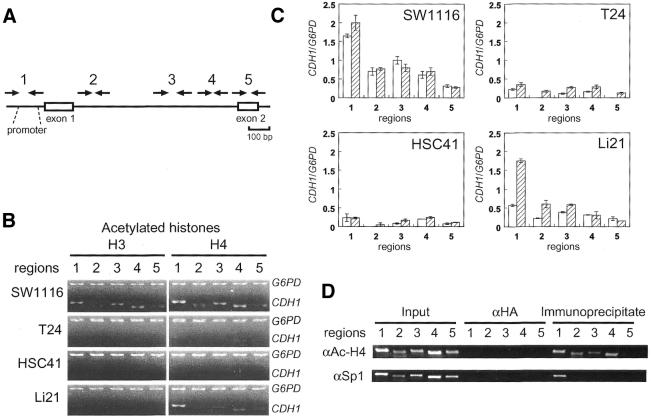
Figure 3.
Differential deacetylation of histones H3 and H4 in the CpG island. (A) Positions of PCR primers used for ChIP analysis. (B) Deacetylation of histones H3 and H4 in the CpG island regions of the CDH1 gene in SW1116, T24, HSC41 and Li21 cells were detected by duplex PCR analysis of immunoprecipitated DNA. The promoter region of the G6PD gene was analyzed for the association with hyperacetylated histones. Input controls are identical to those shown in Figure 4B. (C) Acetylation levels of histone H3 and H4 of the regions 1–5 relative to those of a region within the CpG island of the G6PD gene are shown as open and hatched bars, respectively. Error bars represent the standard deviations of independent duplicate experiments. SW1116, HSC41 and Li21 were established from male patients, and T24 was established from a female patient. The CDH1/G6PD ratio is not compensated for on the basis of the number of X chromosomes in each cell line. (D) Global histone acetylation and localized Sp1 protein binding in the CpG island of SW1116 cell. SW1116 cell lysate was divided into two fractions and each fraction was used for ChIP using anti-acetyl-histone H4 or anti-Sp1 protein antibodies (αAc-H4 or αSp1, respectively).