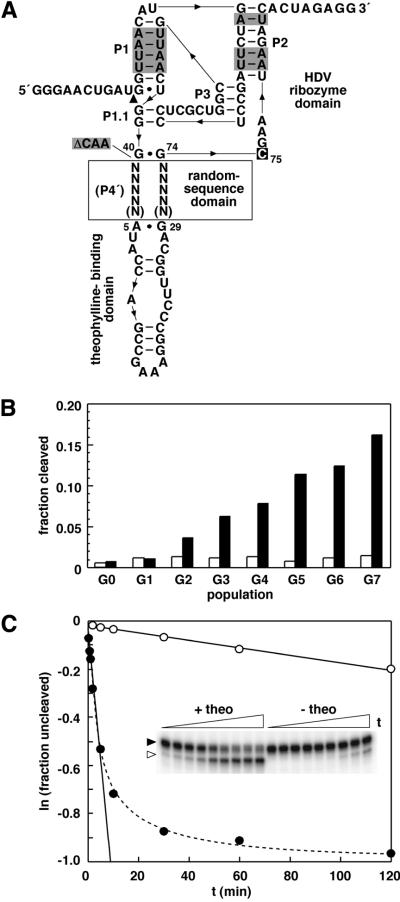
Figure 1.
Isolation and characterization of theophylline-dependent HDV ribozymes. (A) Design of initial populations for in vitro selection. The genomic HDV ribozyme was modified to include random-sequence domains of either 9 or 10 total nucleotides (N) and a theophylline-binding domain in place of P4 (P4′). Other modifications to the ribozyme include the replacement of G-C base pairs with A-U base pairs (shaded) and the inclusion of both 5′- and 3′-terminal extensions to enable selection and facilitate manipulation by RT–PCR. Additionally, 3 nt at positions 41–43 of the genomic HDV ribozyme were deleted (ΔCAA; shaded) as their absence or presence has little effect on ribozyme activity. C75 (boxed) is the nucleotide that functions at the active site of the catalyst as a general acid/base. (B) Progress of selection for theophylline-dependent HDV catalysts. The self-cleavage activity of the initial population (G0) and each subsequent population derived by in vitro selection for theophylline-dependent function is shown for reactions in the absence (open bars) or presence (filled bars) of theophylline under selection conditions incubated for 2 min. (C) Self-cleavage activity of an allosteric HDV ribozyme identified from the final population (G7). The inset depicts the precursor (filled arrowhead) and 3′-cleavage product (open arrowhead) of the theophylline-dependent HDV catalyst containing cm+theo6 as separated by denaturing PAGE following reaction under selection conditions in the absence or presence of theophylline for various lengths of time. The observed rate constants in the absence (open circles) or presence (filled circles) of theophylline were derived by plotting the natural logarithm of the fraction of uncleaved RNA versus time, and establishing the negative slope of the resulting lines (solid lines). The biphasic kinetic profile (dashed line) exhibited by the catalyst in the presence of theophylline indicates that ∼50% of the molecules might be irreversibly misfolded or only slowly convert to the active conformation.