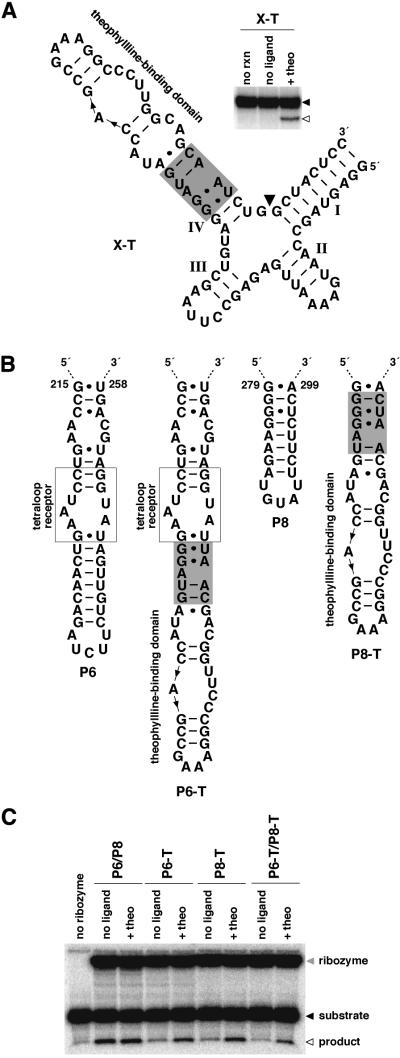
Figure 4.
Identification of effector-dependent sites in X motif and Tetrahymena group I ribozymes. (A) An allosteric X motif ribozyme. An X motif ribozyme was modified by replacing all but two base pairs of stem IV with cm+theo6 and a theophylline-binding domain (X-T). The inset depicts the activity of the allosteric ribozyme. Shown is the uncleaved ribozyme (filled arrowhead) and 5′-cleavage product (open arrowhead) separated by denaturing PAGE following reaction in the absence (no ligand) or presence of theophylline (+theo) at 37°C for 45 min. Unreacted ribozyme is indicated (no rxn). (B) Design of allosteric Tetrahymena group I ribozymes. Group I ribozymes correspond to the L-21 NheI RNA which lacks the P9.1 and P9.2 extensions, and contain CGAAA in place of nucleotides 322–326 in P9. The ribozyme was modified by replacing a portion of P6 or P8 with cm+theo6 and a theophylline-binding domain (P6-T or P8-T, respectively). Alternatively, the ribozyme was modified at both positions (P6-T/P8-T). (C) Activities of unmodified and allosteric Tetrahymena group I ribozymes. Shown is ribozyme (shaded arrowhead), substrate (filled arrowhead) and 5′-cleavage product (open arrowhead) resulting from ribozyme catalysis of the reverse of the second step of splicing (3′-exon ligation). Products were separated by denaturing PAGE following reaction in the absence (no ligand) or presence of theophylline (+theo) at 23°C for 30 min. Unmodified ribozyme is labeled P6/P8. Reaction containing substrate alone is indicated (no ribozyme).