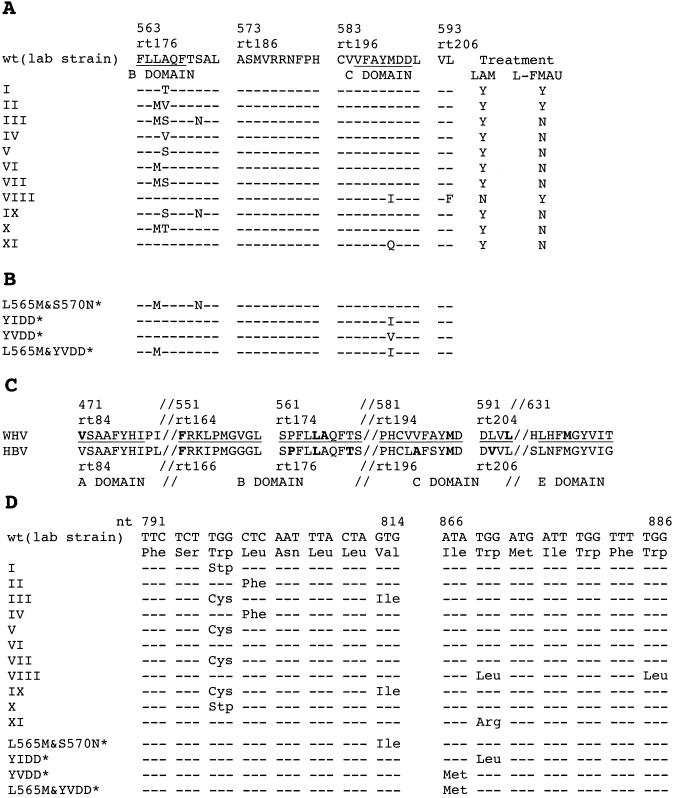
FIG. 1.
Mutations in the polymerase active site. (A) Eleven WHV variants (types I to XI) with mutations in the polymerase active site were detected in the sera of lamivudine- and/or l-FMAU-treated woodchucks. Two (VIII and XI) have mutations in the C region; the remainder have mutations only in the B region. The amino acid sequence of a cloned wild-type WHV polymerase (12, 27) is shown at the top. The columns to the right indicate which drug treatment was associated with the emergence of a variant. (B) Sequence of polymerase mutants of WHV produced for phenotypic analyses. These particular variants have not been found, so far, to occur in drug-treated woodchucks. The YIDD, YVDD, and L565M+YVDD mutants were analogous to HBV mutations that arise in patients subjected to lamivudine therapy. (C) Comparison of amino acid sequences in conserved regions of the HBV and WHV genomes. Amino acids known to be mutated in drug-resistant variants are shown in boldface. (D) Predicted mutations in the overlapping envelope protein due to drug resistance mutations of the WHV polymerase. Amino acid changes in the envelope were predicted for all but the type VI mutant. Among these, a tryptophan-to-stop codon change was predicted for type I and X mutants. In panels A and C, amino acid positions in the WHV polymerase ORF are numbered from the first AUG of polymerase and from the beginning of the reverse transcriptase region (23); only the latter convention is used for HBV.