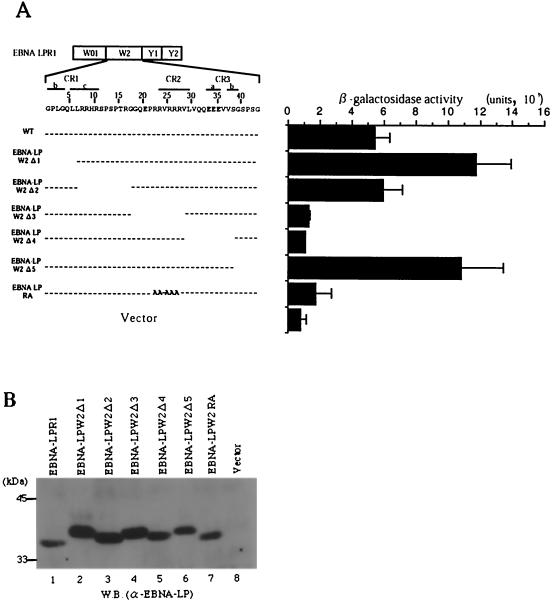
FIG. 5.
Fine mapping of the region responsible for self-interaction of EBNA-LP. (A, left) Line 1 is a schematic representation of the exon structures of EBNA-LPR1. Line 2 shows the predicted amino acid sequence of the EBNA-LP W2 domain. Conserved regions defined by Peng et al. (32) are indicated as lines with their designation given above. Internal deletion and amino acid substitution mutants of the EBNA-LPW2 domain are shown below the amino acid sequence of W2. (A, right) β-Galactosidase activity detected for each construct in the yeast two-hybrid system, determined by liquid assay procedures as described in the legend to Fig. 1. (B) Immunoblot of electrophoretically separated lysates from yeasts transfected with expression plasmids for wild-type and mutant EBNA-LPR1 proteins shown in panel A. Yeast protein lysates were prepared as described in Materials and Methods and subjected to electrophoresis on a denaturing gel, transferred to a nitrocellulose sheet, and reacted with the anti-EBNA-LP monoclonal antibody. Positions of molecular mass markers are on the left. W.B., Western blot.