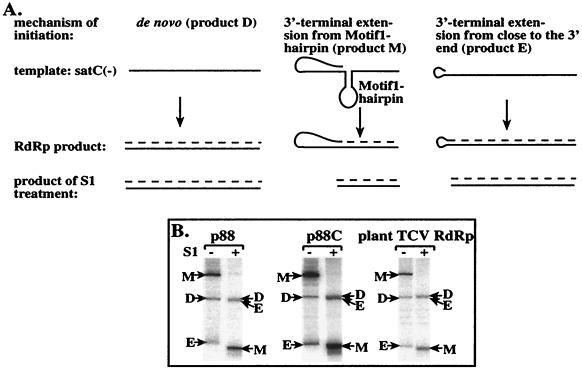
FIG. 3.
Comparison of the RdRp products obtained with the recombinant p88 and p88C and with the TCV RdRp purified from plants using satC(−) as the template. (A) Schematic representation of the three major products generated by the plant TCV RdRp in vitro using satC(−) templates. The mechanism of the formation of these products is explained schematically based on data from Song and Simon (39, 41). Solid lines represent the template, while the broken line depicts the cRNA product (labeled during the RdRp reaction). S1 nuclease treatment removes the single-stranded regions of the RdRp products under the conditions used (28, 41). The names of the products reflect their generation: product D, de novo initiation; product M, motif 1 hairpin-mediated primer extension; product E, 3"-end-mediated primer extension. (B) Representative denaturing gel analyses of radiolabeled RNA products synthesized by in vitro transcription with the recombinant p88 and p88C and the plant TCV RdRp. The − and + signs above the lanes indicate untreated and S1 nuclease-treated samples, respectively, prior to PAGE analysis. Products M, D, and E are marked with arrows. Note that product E runs aberrantly (i.e., much faster) under these conditions in the untreated samples due to the highly stable hairpin structure of this product (see panel A and also reference 41). Also, S1-treated product E migrates close to product D under the conditions used. The exposure time was different for each set of RdRps due to the different activities of the RdRp preparations. The amount of p88C applied in the RdRp assay was 40% of that of p88, based on Coomassie-stained SDS-PAGE gels (not shown).